predict
Classify observations using naive Bayes classifier
Description
[ also returns the
Posterior Probability (label,Posterior,Cost]
= predict(Mdl,X)Posterior) and predicted
(expected) Misclassification Cost (Cost) corresponding to
the observations (rows) in Mdl.X. For each observation in
X, the predicted class label corresponds to the minimum
expected classification cost among all classes.
Examples
Load the fisheriris data set. Create X as a numeric matrix that contains four measurements for 150 irises. Create Y as a cell array of character vectors that contains the corresponding iris species.
load fisheriris X = meas; Y = species; rng('default') % for reproducibility
Randomly partition observations into a training set and a test set with stratification, using the class information in Y. Specify a 30% holdout sample for testing.
cv = cvpartition(Y,'HoldOut',0.30);Extract the training and test indices.
trainInds = training(cv); testInds = test(cv);
Specify the training and test data sets.
XTrain = X(trainInds,:); YTrain = Y(trainInds); XTest = X(testInds,:); YTest = Y(testInds);
Train a naive Bayes classifier using the predictors XTrain and class labels YTrain. A recommended practice is to specify the class names. fitcnb assumes that each predictor is conditionally and normally distributed.
Mdl = fitcnb(XTrain,YTrain,'ClassNames',{'setosa','versicolor','virginica'})
Mdl =
ClassificationNaiveBayes
ResponseName: 'Y'
CategoricalPredictors: []
ClassNames: {'setosa' 'versicolor' 'virginica'}
ScoreTransform: 'none'
NumObservations: 105
DistributionNames: {'normal' 'normal' 'normal' 'normal'}
DistributionParameters: {3×4 cell}
Properties, Methods
Mdl is a trained ClassificationNaiveBayes classifier.
Predict the test sample labels.
idx = randsample(sum(testInds),10); label = predict(Mdl,XTest);
Display the results for a random set of 10 observations in the test sample.
table(YTest(idx),label(idx),'VariableNames',... {'TrueLabel','PredictedLabel'})
ans=10×2 table
TrueLabel PredictedLabel
______________ ______________
{'virginica' } {'virginica' }
{'versicolor'} {'versicolor'}
{'versicolor'} {'versicolor'}
{'virginica' } {'virginica' }
{'setosa' } {'setosa' }
{'virginica' } {'virginica' }
{'setosa' } {'setosa' }
{'versicolor'} {'versicolor'}
{'versicolor'} {'virginica' }
{'versicolor'} {'versicolor'}
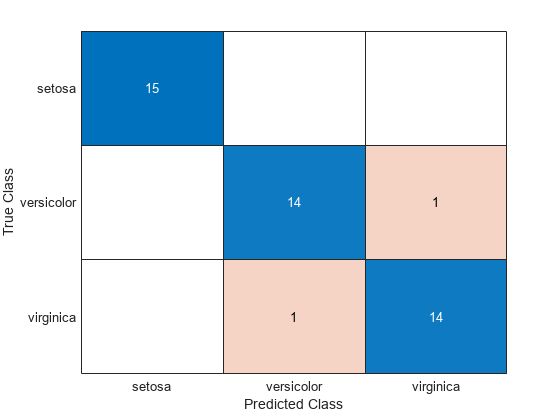
Create a confusion chart from the true labels YTest and the predicted labels label.
cm = confusionchart(YTest,label);
Estimate posterior probabilities and misclassification costs for new observations using a naive Bayes classifier. Classify new observations using a memory-efficient pretrained classifier.
Load the fisheriris data set. Create X as a numeric matrix that contains four measurements for 150 irises. Create Y as a cell array of character vectors that contains the corresponding iris species.
load fisheriris X = meas; Y = species; rng('default') % for reproducibility
Partition the data set into two sets: one contains the training set, and the other contains new, unobserved data. Reserve 10 observations for the new data set.
n = size(X,1); newInds = randsample(n,10); inds = ~ismember(1:n,newInds); XNew = X(newInds,:); YNew = Y(newInds);
Train a naive Bayes classifier using the predictors X and class labels Y. A recommended practice is to specify the class names. fitcnb assumes that each predictor is conditionally and normally distributed.
Mdl = fitcnb(X(inds,:),Y(inds),... 'ClassNames',{'setosa','versicolor','virginica'});
Mdl is a trained ClassificationNaiveBayes classifier.
Conserve memory by reducing the size of the trained naive Bayes classifier.
CMdl = compact(Mdl); whos('Mdl','CMdl')
Name Size Bytes Class Attributes CMdl 1x1 5534 classreg.learning.classif.CompactClassificationNaiveBayes Mdl 1x1 12907 ClassificationNaiveBayes
CMdl is a CompactClassificationNaiveBayes classifier. It uses less memory than Mdl because Mdl stores the data.
Display the class names of CMdl using dot notation.
CMdl.ClassNames
ans = 3×1 cell
{'setosa' }
{'versicolor'}
{'virginica' }
Predict the labels. Estimate the posterior probabilities and expected class misclassification costs.
[labels,PostProbs,MisClassCost] = predict(CMdl,XNew);
Compare the true labels with the predicted labels.
table(YNew,labels,PostProbs,MisClassCost,'VariableNames',... {'TrueLabels','PredictedLabels',... 'PosteriorProbabilities','MisclassificationCosts'})
ans=10×4 table
TrueLabels PredictedLabels PosteriorProbabilities MisclassificationCosts
______________ _______________ _________________________________________ ______________________________________
{'virginica' } {'virginica' } 4.0832e-268 4.6422e-09 1 1 1 4.6422e-09
{'setosa' } {'setosa' } 1 3.0706e-18 4.6719e-25 3.0706e-18 1 1
{'virginica' } {'virginica' } 1.0007e-246 5.8758e-10 1 1 1 5.8758e-10
{'versicolor'} {'versicolor'} 1.2022e-61 0.99995 4.9859e-05 1 4.9859e-05 0.99995
{'virginica' } {'virginica' } 2.687e-226 1.7905e-08 1 1 1 1.7905e-08
{'versicolor'} {'versicolor'} 3.3431e-76 0.99971 0.00028983 1 0.00028983 0.99971
{'virginica' } {'virginica' } 4.05e-166 0.0028527 0.99715 1 0.99715 0.0028527
{'setosa' } {'setosa' } 1 1.1272e-14 2.0308e-23 1.1272e-14 1 1
{'virginica' } {'virginica' } 1.3292e-228 8.3604e-10 1 1 1 8.3604e-10
{'setosa' } {'setosa' } 1 4.5023e-17 2.1724e-24 4.5023e-17 1 1
PostProbs and MisClassCost are 10-by-3 numeric matrices, where each row corresponds to a new observation and each column corresponds to a class. The order of the columns corresponds to the order of CMdl.ClassNames.
Load the fisheriris data set. Create X as a numeric matrix that contains petal length and width measurements for 150 irises. Create Y as a cell array of character vectors that contains the corresponding iris species.
load fisheriris
X = meas(:,3:4);
Y = species;Train a naive Bayes classifier using the predictors X and class labels Y. A recommended practice is to specify the class names. fitcnb assumes that each predictor is conditionally and normally distributed.
Mdl = fitcnb(X,Y,'ClassNames',{'setosa','versicolor','virginica'});
Mdl is a trained ClassificationNaiveBayes classifier.
Define a grid of values in the observed predictor space.
xMax = max(X); xMin = min(X); h = 0.01; [x1Grid,x2Grid] = meshgrid(xMin(1):h:xMax(1),xMin(2):h:xMax(2));
Predict the posterior probabilities for each instance in the grid.
[~,PosteriorRegion] = predict(Mdl,[x1Grid(:),x2Grid(:)]);
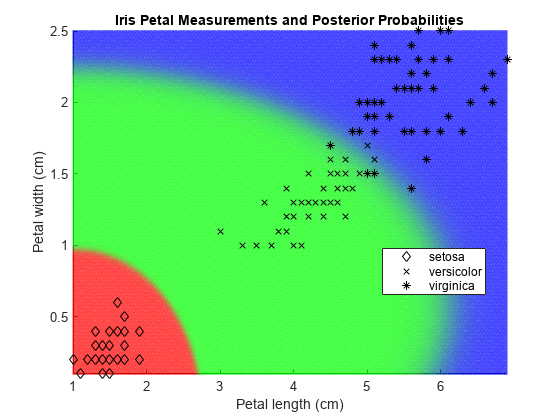
Plot the posterior probability regions and the training data.
h = scatter(x1Grid(:),x2Grid(:),1,PosteriorRegion); h.MarkerEdgeAlpha = 0.3;
Plot the data.
hold on gh = gscatter(X(:,1),X(:,2),Y,'k','dx*'); title 'Iris Petal Measurements and Posterior Probabilities'; xlabel 'Petal length (cm)'; ylabel 'Petal width (cm)'; axis tight legend(gh,'Location','Best') hold off
Input Arguments
Naive Bayes classification model, specified as a ClassificationNaiveBayes model object or CompactClassificationNaiveBayes model object returned by fitcnb or compact,
respectively.
Predictor data to be classified, specified as a numeric matrix or table.
Each row of X corresponds to one observation, and
each column corresponds to one variable.
For a numeric matrix:
The variables that make up the columns of
Xmust have the same order as the predictor variables that trainedMdl.If you train
Mdlusing a table (for example,Tbl), thenXcan be a numeric matrix ifTblcontains only numeric predictor variables. To treat numeric predictors inTblas categorical during training, identify categorical predictors using the'CategoricalPredictors'name-value pair argument offitcnb. IfTblcontains heterogeneous predictor variables (for example, numeric and categorical data types) andXis a numeric matrix, thenpredictthrows an error.
For a table:
predictdoes not support multicolumn variables or cell arrays other than cell arrays of character vectors.If you train
Mdlusing a table (for example,Tbl), then all predictor variables inXmust have the same variable names and data types as the variables that trainedMdl(stored inMdl.PredictorNames). However, the column order ofXdoes not need to correspond to the column order ofTbl.TblandXcan contain additional variables (response variables, observation weights, and so on), butpredictignores them.If you train
Mdlusing a numeric matrix, then the predictor names inMdl.PredictorNamesmust be the same as the corresponding predictor variable names inX. To specify predictor names during training, use the 'PredictorNames' name-value pair argument offitcnb. All predictor variables inXmust be numeric vectors.Xcan contain additional variables (response variables, observation weights, and so on), butpredictignores them.
Data Types: table | double | single
Notes:
If
Mdl.DistributionNamesis'mn', then the software returnsNaNs corresponding to rows ofXthat contain at least oneNaN.If
Mdl.DistributionNamesis not'mn', then the software ignoresNaNvalues when estimating misclassification costs and posterior probabilities. Specifically, the software computes the conditional density of the predictors given the class by leaving out the factors corresponding to missing predictor values.For predictor distribution specified as
'mvmn', ifXcontains levels that are not represented in the training data (that is, not inMdl.CategoricalLevelsfor that predictor), then the conditional density of the predictors given the class is 0. For those observations, the software returns the corresponding value ofPosterioras aNaN. The software determines the class label for such observations using the class prior probability stored inMdl.Prior.
Output Arguments
Predicted class labels, returned as a categorical vector, character array, logical or numeric vector, or cell array of character vectors.
The predicted class labels have the following:
Same data type as the observed class labels (
Mdl.Y). (The software treats string arrays as cell arrays of character vectors.)Length equal to the number of rows of
Mdl.X.Class yielding the lowest expected misclassification cost (
Cost).
Class Posterior Probability, returned as a numeric matrix.
Posterior has rows equal to the number of rows of
Mdl.X and columns equal to the number of distinct classes in the
training data (size(Mdl.ClassNames,1)).
Posterior(j,k) is the predicted posterior probability of class
k (in class Mdl.ClassNames(k)) given the
observation in row j of Mdl.X.
Expected Misclassification Cost, returned as a numeric matrix.
Cost has rows equal to the number of rows of
Mdl.X and columns equal to the number of distinct classes in the
training data (size(Mdl.ClassNames,1)).
Cost(j,k) is the expected misclassification cost of the observation in row
j of Mdl.X predicted into class
k (in class Mdl.ClassNames(k)).
More About
A misclassification cost is the relative severity of a classifier labeling an observation into the wrong class.
Two types of misclassification cost exist: true and expected. Let K be the number of classes.
True misclassification cost — A K-by-K matrix, where element (i,j) indicates the cost of classifying an observation into class j if its true class is i. The software stores the misclassification cost in the property
Mdl.Cost, and uses it in computations. By default,Mdl.Cost(i,j)= 1 ifi≠j, andMdl.Cost(i,j)= 0 ifi=j. In other words, the cost is0for correct classification and1for any incorrect classification.Expected misclassification cost — A K-dimensional vector, where element k is the weighted average cost of classifying an observation into class k, weighted by the class posterior probabilities.
In other words, the software classifies observations into the class with the lowest expected misclassification cost.
The posterior probability is the probability that an observation belongs in a particular class, given the data.
For naive Bayes, the posterior probability that a classification is k for a given observation (x1,...,xP) is
where:
is the conditional joint density of the predictors given they are in class k.
Mdl.DistributionNamesstores the distribution names of the predictors.π(Y = k) is the class prior probability distribution.
Mdl.Priorstores the prior distribution.is the joint density of the predictors. The classes are discrete, so
The prior probability of a class is the assumed relative frequency with which observations from that class occur in a population.
Alternative Functionality
Simulink Block
To integrate the prediction of a naive Bayes classification model into Simulink®, you can use the ClassificationNaiveBayes Predict block in the Statistics and Machine Learning Toolbox™ library or a MATLAB® Function block with the predict function. For
examples, see Predict Class Labels Using ClassificationNaiveBayes Predict Block and Predict Class Labels Using MATLAB Function Block.
When deciding which approach to use, consider the following:
If you use the Statistics and Machine Learning Toolbox library block, you can use the Fixed-Point Tool (Fixed-Point Designer) to convert a floating-point model to fixed point.
Support for variable-size arrays must be enabled for a MATLAB Function block with the
predictfunction.If you use a MATLAB Function block, you can use MATLAB functions for preprocessing or post-processing before or after predictions in the same MATLAB Function block.
Extended Capabilities
This function fully supports tall arrays. You can use models trained on either in-memory or tall data with this function.
For more information, see Tall Arrays.
Usage notes and limitations:
Use
saveLearnerForCoder,loadLearnerForCoder, andcodegen(MATLAB Coder) to generate code for thepredictfunction. Save a trained model by usingsaveLearnerForCoder. Define an entry-point function that loads the saved model by usingloadLearnerForCoderand calls thepredictfunction. Then usecodegento generate code for the entry-point function.To generate single-precision C/C++ code for
predict, specifyDataType="single"when you call theloadLearnerForCoderfunction.This table contains notes about the arguments of
predict. Arguments not included in this table are fully supported.Argument Notes and Limitations MdlFor the usage notes and limitations of the model object, see Code Generation of the
CompactClassificationNaiveBayesobject.XXmust be a single-precision or double-precision matrix or a table containing numeric variables, categorical variables, or both.The number of rows, or observations, in
Xcan be a variable size, but the number of columns inXmust be fixed.If you want to specify
Xas a table, then your model must be trained using a table, and your entry-point function for prediction must do the following:Accept data as arrays.
Create a table from the data input arguments and specify the variable names in the table.
Pass the table to
predict.
For an example of this table workflow, see Generate Code to Classify Data in Table. For more information on using tables in code generation, see Code Generation for Tables (MATLAB Coder) and Table Limitations for Code Generation (MATLAB Coder).
For more information, see Introduction to Code Generation.
Version History
Introduced in R2014b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)