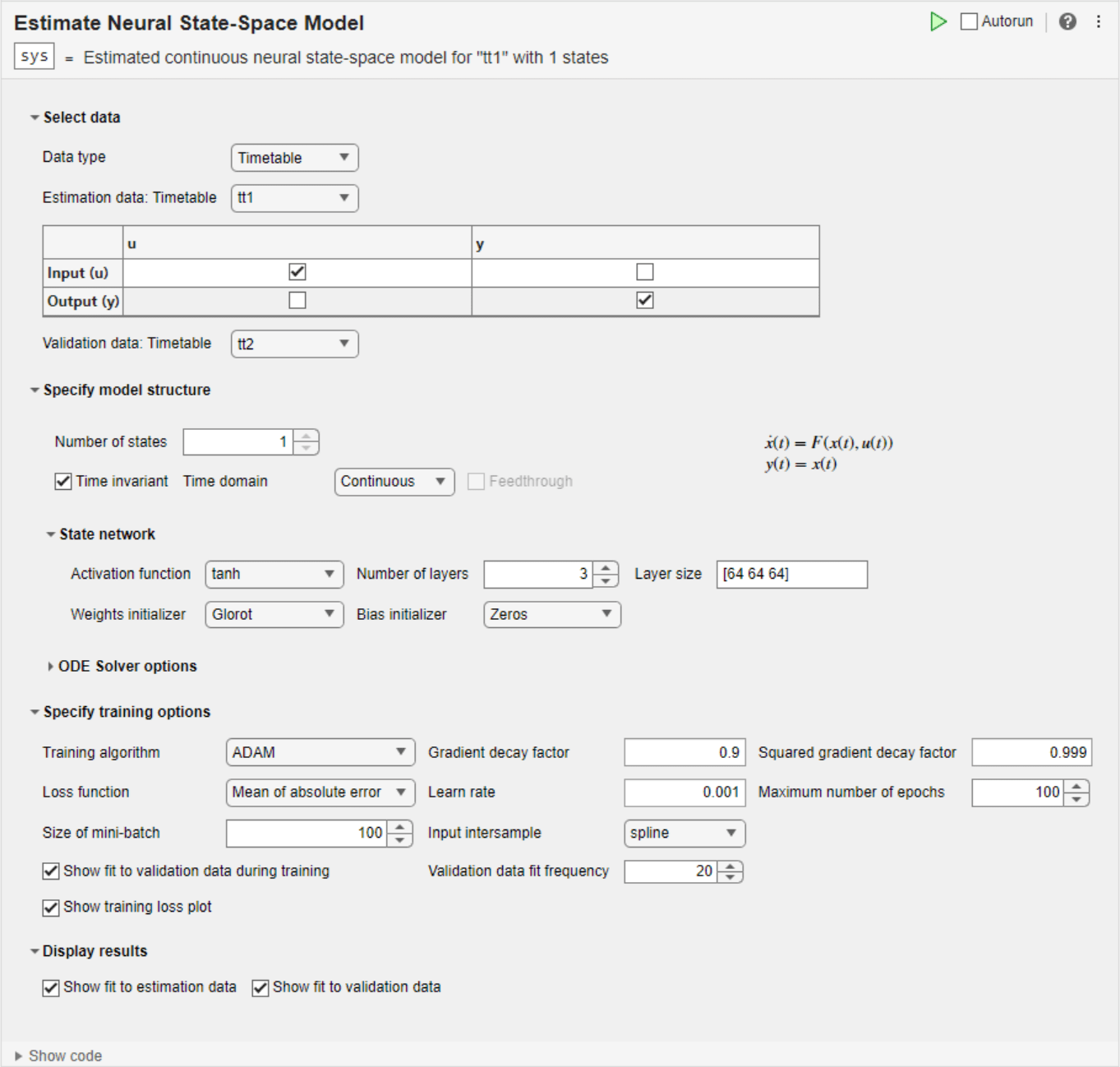
Estimate Neural State-Space Model
Description
The Estimate Neural State-Space Model task lets you interactively estimate and validate a neural state-space model, using time-domain data. You can define and vary the structure and the parameters of the networks and the solver. The task automatically generates MATLAB® code for your live script. For more information about Live Editor tasks, see Add Interactive Tasks to a Live Script. For more information about state-space estimation, see What Are State-Space Models?
The Estimate Neural State-Space Model task is independent of the more general System Identification app. Use the System Identification app when you want to compute and compare estimates for multiple model structures.
To get started, load experiment data that contains input and output data into your MATLAB workspace and then import that data into the task. Then specify a model structure to estimate. The task gives you controls and plots that help you experiment with different model parameters and compare how well the output of each model fits the measurements.
Related Functions
The code that Estimate Neural State-Space Model generates uses the following functions and objects.
The task estimates an idNeuralStateSpace
state-space model.
Open the Task
To add the Estimate Neural State-Space Model task to a live script in the MATLAB Editor:
On the Live Editor tab, select Task > Estimate Neural State-Space Model.
In a code block in your script, type a relevant keyword, such as
neuralstatespaceornlssest. SelectEstimate State-Space Modelfrom the suggested command completions.
Examples
Use the Estimate Neural State-Space Model Live Editor Task to estimate a neural state-space model and compare the model output with the measurement data.
Open this example to see a preconfigured script containing the task.
Generate Data
For this example, generate data by simulating a first-order linear system. First, fix the random generator seed to guarantee reproducibility.
rng(0)
Create a first-order discrete dynamical system in tf form with one input and one output, convert it to discrete time using a sample time of 0.1 sec, and use ss to obtain a state-space realization.
Ts = 0.1; sys = ss(c2d(tf(1,[1 1]),Ts));
The identification of a neural state-space system requires you to have measurement of the system states. Therefore, transform the state-space coordinates so that the output is equal to the state. Alternatively you can augment the output equation to include the state among the measured signals.
sys.b = sys.b*sys.c; sys.c = 1;
In general, it is good practice to use multiple experiments, each containing a different trajectory, as doing so is more likely to yield a better coverage of the state-input space. Furthermore, using long trajectories tends to reduce both the accuracy and efficiency of the estimation. However, for this example, use a single trajectory for estimation.
Define a time vector and a random input sequence for estimation (training).
te = 0:Ts:10; ue = randn(length(te),size(sys.B,2));
Generate an output response to the random input sequence by simulating the system from a zero initial condition. The first (vertical) dimension in ye must be time and the second (horizontal) dimension must be the specific output in the output vector signal.
ye = lsim(sys,ue,te,zeros(size(sys.B,1),1));
Define a shorter time vector and a random input sequence for validation.
tv = 0:Ts:1; uv = randn(length(tv),size(sys.B,2));
Generate an output response to the random input sequence by simulating the system, from a zero initial condition.
yv = lsim(sys,uv,tv,zeros(size(sys.B,1),1));
Import Data into the Task
In the Select data section, set Data Type to Numeric, Sample Time to 0.01, Estimation Data: Input (u) to ue, Estimation Data: Output (y) to ye, Validation Data: Input (u) to uv, and Validation Data: Output (y) to yv.

Specify Model Structure and State Network
In the Specify model structure section, set the Number of states to 1 and select the discrete-time domain. In the State network section, set the Number of layers to 1 and specify Layer size as 16. Leave the other options unchanged.

Note that since for this example the output is equal to the state, there is no Output network section. Since the latent dimension is not specified, there are no Encoder network and Decoder network sections.
Examine Training and Display Options
In the Specify training options section, the Training algorithm is set to ADAM, with a Learn rate of 0.001. The number of epochs is set to 100, and so is the mini-batch size. For more information on these options, see nssTrainingOptions.
In the Display results section, both the Show fit to estimation data and (since you have specified validation data) the Show fit to validation data are selected.

Execute Live Task
Execute the task from the Live Editor tab using Run. During training, a plot displays the training losses of the state and output networks.

Generating estimation report...done.


After training, two plots displays the model fit on the estimation and validation data.
Generate Code
To display the code that the task generates, click ![]() (Show code) at the bottom of the parameter section. The code that you see reflects the current parameter configuration of the task.
(Show code) at the bottom of the parameter section. The code that you see reflects the current parameter configuration of the task.



Related Examples
Parameters
Select data
The task accepts numeric measurement values that are uniformly sampled in time.
Input and output signals can contain multiple channels. Data can be packaged as numeric
arrays, in an iddata object, or in a timetable object. For multiexperiment data, numeric and timetable data can
be packaged as cell arrays. For cell arrays of timetables, all timetables must contain
the same variable names. Data objects handle multiexperiment data internally.
The data type you choose determines whether you must specify additional parameters.
Numeric— Specify Sample Time and Start Time in the time unit that you select. Additionally, you need to specify different workspace variables containing the input and output signals to be used for estimation and (if available) validation.Timetable— Specify no additional parameters because the timetable already contains the input and output signals and sampling information.iddata object— Specify no additional parameters because theiddataobject already contains the input and output signals and sampling information.
Sample time at which estimation and (if available) validation data are collected,
specified as a positive scalar, in the unit specified by the following time unit drop
down list. You can specify a sample time only when Data Type is
Numeric.
Time unit for the Sample time and Start time parameters. You can specify one of the following units:
nanosecondsmicrosecondsmillisecondssecondsminuteshoursdaysweeksmonthsyears
You can specify a sample time only when Data Type is
Numeric.
Start time for the estimation and (if available) validation data, specified as a
nonnegative scalar, in the unit specified by the preceding time unit drop down list.
This value is relevant only if you deselect the Time invariant
checkbox. You can specify a start time only when Data Type is
Numeric.
Name of the input data variable used for estimation, selected from the MATLAB workspace choices. Use this parameter, along with Estimation
data: Output (y), when Data Type is
Numeric.
Name of the output data variable used for estimation, selected from the MATLAB workspace choices. Use this parameter, along with Estimation
data: Input (u), when Data Type is
Numeric.
Select the timetable object variable name from the MATLAB workspace choices. If you use a use a cell arrays of timetables, all
timetables must contain the same variable names. Use this parameter when Data
type is Timetable.
Select the iddata object variable name from the MATLAB workspace choices. Use this parameter when Data type
is iddata object.
Name of the input data variable used for validation, selected from the MATLAB workspace choices. Use this parameter, along with Validation
data: Output (y), when Data Type is
Numeric.
Name of the output data variable used for validation, selected from the MATLAB workspace choices. Use this parameter, along with Validation
data: Input (y), when Data Type is
Numeric.
Select the timetable object variable name from the MATLAB workspace choices. The timetables containing the validation data must have
the same variable names as the ones in the timetables selected for estimation in
Estimation data: Timetable. Use this parameter when
Data type is Timetable. Specifying
validation data is optional but recommended.
Select the iddata object variable name from the MATLAB workspace choices. Use this parameter when Data type
is iddata object. Specifying validation data is optional but
recommended.
Specify model structure
Number of states in the model to estimate. It must be less than or equal to the
number of outputs in the data. For more information, see idNeuralStateSpace.
Dimension of the internal (latent) state. When this option is left blank (default),
there is no encoder or decoder in the model. To add an encoder or decoder to your model,
specify this option as a finite positive integer. For more information, see the
LatentDim property of idNeuralStateSpace.
Deselect this option to estimate a model in which the state equation explicitly
depends on time, other than states and inputs. When this option is left selected
(default), the state equation depends explicitly only on the current state and input
vectors. For more information, see the isTimeInvariant property of
idNeuralStateSpace.
Select a continuous-time or discrete-time model.
Select this option to estimate a model in which the output equation explicitly
depends on the input vector. When this option is left unselected (default), the output
equation does not depend explicitly on the input vector. For more information, see the
HasFeedthrough property of idNeuralStateSpace.
State network
You can specify one of the following as the activation function for all hidden
layers of the state network: tanh,
sigmoid, relu,
leakyRelu, clippedRelu,
elu, gelu,
swish, softplus,
scaling, or softmax. All of
these are available in Deep Learning Toolbox™.
Also, you can now choose to not use an activation function by specifying the
activation function as none.
For more information, see createMLPNetwork.
Number of hidden layers of the state network, specified as a nonnegative integer. It
must be equal to the number of elements of the vector you specify in Layer
size in the State network section. If you specify
0, the state network has no hidden layer, and therefore expresses a
linear function.
Size of the hidden layers for the state network, specified as a vector of positive
integers. Each number specifies the number of neurons (network nodes) for each hidden
layer (each layer is fully-connected). For example, [10 20 8]
specifies a network with three hidden layers, the first (after the network input) having
10 neurons, the second having 20 neurons, and
the last (before the network output), having 8 neurons. Note that the
output layer is also fully-connected, and you cannot change its size.
The number of elements in Layer size must be equal to the value specified in Number of layers in the State network section.
Weights initializer method for all the hidden layers of the state network. You can specify one of the following:
glorot— uses the Glorot method (default).he— uses the He method.orthogonal— uses the orthogonal method.narrow-normal— uses the narrow-normal method.zeros— initializes all weights to zero.ones— initializes all weights to one.
Bias initializer method for all the hidden layers of the state network. You can specify one of the following:
zeros— initializes all biases to zero (default).ones— initializes all biases to one.narrow-normal— uses the narrow-normal method.
Output network
You can specify one of the following as the activation function for all hidden
layers of the output network: tanh,
sigmoid, relu,
leakyRelu, clippedRelu,
elu, gelu,
swish, softplus,
scaling, or softmax. All of
these are available in Deep Learning Toolbox.
Also, you can now choose to not use an activation function by specifying the
activation function as none.
For more information, see createMLPNetwork.
Number of hidden layers of the output network, specified as a nonnegative integer.
It must be equal to the number of elements of the vector you specify in Layer
size in the Output network section. If you specify
0, the output network has no hidden layer, and therefore expresses
a linear function.
Size of the hidden layers for the output network, specified as a vector of positive
integers. Each number specifies the number of neurons (network nodes) for each hidden
layer (each layer is fully-connected). For example, [10 20 8]
specifies a network with three hidden layers, the first (after the network input) having
10 neurons, the second having 20 neurons, and
the last (before the network output), having 8 neurons. Note that the
output layer is also fully-connected, and you cannot change its size.
The number of elements Layer size must be equal to the value specified in Number of layers in the Output network section.
Weights initializer method for all the hidden layers of the output network. You can specify one of the following:
glorot— uses the Glorot method (default).he— uses the He method.orthogonal— uses the orthogonal method.narrow-normal— uses the narrow-normal method.zeros— initializes all weights to zero.ones— initializes all weights to one.
Bias initializer method for all the hidden layers of the output network. You can specify one of the following:
zeros— initializes all biases to zero (default).ones— initializes all biases to one.narrow-normal— uses the narrow-normal method.
Encoder network
You can specify one of the following as the activation function for all hidden
layers of the encoder network: tanh,
sigmoid, relu,
leakyRelu, clippedRelu,
elu, gelu,
swish, softplus,
scaling, or softmax. All of
these are available in Deep Learning Toolbox.
Also, you can now choose to not use an activation function by specifying the
activation function as none.
For more information, see createMLPNetwork.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Number of hidden layers of the encoder network, specified as a nonnegative integer.
It must be equal to the number of elements of the vector you specify in Layer
size in the Encoder network section. If you specify
0, the encoder network has no hidden layer, and therefore expresses
a linear function.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Size of the hidden layers for the encoder network, specified as a vector of positive
integers. Each number specifies the number of neurons (network nodes) for each hidden
layer (each layer is fully-connected). For example, [10 20 8]
specifies a network with three hidden layers, the first (after the network input) having
10 neurons, the second having 20 neurons, and
the last (before the network output), having 8 neurons. Note that the
output layer is also fully-connected, and you cannot change its size.
The number of elements in Layer size must be equal to the value specified in Number of layers in the Encoder network section.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Weights initializer method for all the hidden layers of the encoder network. You can specify one of the following:
glorot— uses the Glorot method (default).he— uses the He method.orthogonal— uses the orthogonal method.narrow-normal— uses the narrow-normal method.zeros— initializes all weights to zero.ones— initializes all weights to one.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Bias initializer method for all the hidden layers of the encoder network. You can specify one of the following:
zeros— initializes all biases to zero (default).ones— initializes all biases to one.narrow-normal— uses the narrow-normal method.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Decoder network
You can specify one of the following as the activation function for all hidden
layers of the decoder network: tanh,
sigmoid, relu,
leakyRelu, clippedRelu,
elu, gelu,
swish, softplus,
scaling, or softmax. All of
these are available in Deep Learning Toolbox.
Also, you can now choose to not use an activation function by specifying the
activation function as none.
For more information, see createMLPNetwork.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Number of hidden layers of the decoder network, specified as a nonnegative integer.
It must be equal to the number of elements of the vector you specify in Layer
size in the Decoder network section. If you specify
0, the decoder network has no hidden layer, and therefore expresses
a linear function.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Size of the hidden layers for the decoder network, specified as a vector of positive
integers. Each number specifies the number of neurons (network nodes) for each hidden
layer (each layer is fully-connected). For example, [10 20 8]
specifies a network with three hidden layers, the first (after the network input) having
10 neurons, the second having 20 neurons, and
the last (before the network output), having 8 neurons. Note that the
output layer is also fully-connected, and you cannot change its size.
The number of elements in Layer size must be equal to the value specified in Number of layers in the Decoder network section.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Weights initializer method for all the hidden layers of the decoder network. You can specify one of the following:
glorot— uses the Glorot method (default).he— uses the He method.orthogonal— uses the orthogonal method.narrow-normal— uses the narrow-normal method.zeros— initializes all weights to zero.ones— initializes all weights to one.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
Bias initializer method for all the hidden layers of the decoder network. You can specify one of the following:
zeros— initializes all biases to zero (default).ones— initializes all biases to one.narrow-normal— uses the narrow-normal method.
Dependencies
To enable this parameter, specify the Latent dimension parameter as a finite positive integer.
ODE Solver options
Initial step size used to simulate the model (when continuous-time). It is specified
as either Auto or a positive scalar. If you specify
Auto, then the solver bases the initial step size on the slope of
the solution at the initial time point.
For more information, see odeset.
Maximum step size used to simulate the model (when continuous-time). It is an upper
bound on the size of any step taken by the solver, and it is specified as either
Auto or a positive scalar. If you specify Auto,
then the value used is one-tenth of the difference between final and initial
time.
For more information, see odeset.
Absolute tolerance used to simulate continuous time models, specified as a positive
scalar. It is the largest allowable absolute error. That is, when the solution
approaches 0, AbsoluteTolerance is the threshold below which you do
not worry about the accuracy of the solution since it is effectively 0.
For more information, see odeset.
Relative tolerance used to simulate the continuous time models, specified as a positive scalar. This tolerance measures the error relative to the magnitude of each solution component. That is, it controls the number of significant digits in a solution (except when is smaller than the absolute tolerance).
For more information, see odeset.
Specify training options
You can specify one of the following:
ADAM— uses the Adam (adaptive moment estimation) algorithm.SGDM— uses the SGDM (stochastic gradient descent with momentum) algorithm.RMSProp— uses the RMSProp (root mean square propagation) algorithm.LBFGS— uses the L-BFGS (limited-memory BFGS) algorithm.
For more information on these algorithms, see the Algorithms section of
trainingOptions (Deep Learning Toolbox).
Decay rate of gradient moving average for the Adam solver, specified as a
nonnegative scalar less than 1. The gradient decay rate is denoted by
β1 in the Adaptive Moment Estimation (Deep Learning Toolbox) section.
The default value works well for most tasks. You can specify a Gradient
decay factor only when Training algorithm is
ADAM.
For more information, see Adaptive Moment Estimation (Deep Learning Toolbox).
Decay rate of squared gradient moving average for the RMSProp solver, specified as a
nonnegative scalar less than 1. The default value is
0.999 for the Adam solver and 0.9 for the
RMSProp solver.
Typical values of the decay rate are 0.9,
0.99, and 0.999, corresponding to averaging
lengths of 10, 100, and 1000
parameter updates, respectively.
You can specify a Squared gradient decay factor only when
Training algorithm is ADAM or
RMSProp.
For more information, see Root Mean Square Propagation (Deep Learning Toolbox).
Contribution of the parameter update step of the previous iteration to the current
iteration of stochastic gradient descent with momentum, specified as a scalar from
0 to 1.
A value of 0 means no contribution from the previous step,
whereas a value of 1 means maximal contribution from the previous
step. The default value works well for most tasks.
You can specify Momentum only when Training
algorithm is SGDM.
For more information, see Stochastic Gradient Descent with Momentum (Deep Learning Toolbox).
Coefficient applied to tune the reconstruction loss of an autoencoder, specified as a nonnegative scalar.
Reconstruction loss measures the difference between the original input
(x) and its reconstruction
(xr) after encoding and decoding. You
calculate this loss as the L2 norm of (x
-
xr) divided by the batch size
(N).
Dependencies
To enable this option, specify the Latent dimension parameter as a finite positive integer.
Constant coefficient applied to the regularization term added to the loss function, specified as a positive scalar.
The loss function with the regularization term is given by:
where t is the time variable, N is the size of the batch, ε is the sum of the reconstruction loss and autoencoder loss, θ is a concatenated vector of weights and biases of the neural network, and λ is the regularization constant that you can tune.
For more information, see Regularized Estimates of Model Parameters.
You can specify one of the following:
Mean of absolute error— uses the mean value of the absolute error.Mean of squared error— uses the mean value of the squared error.
Maximum number of iterations to use for training, specified as a positive integer.
The L-BFGS solver is a full-batch solver, which means that it processes the entire training set in a single iteration.
You can specify Maximum iterations only when Training
algorithm is LBFGS.
Method to find suitable learning rate, specified as one of these values:
"weak-wolfe"— Search for a learning rate that satisfies the weak Wolfe conditions. This method maintains a positive definite approximation of the inverse Hessian matrix."strong-wolfe"— Search for a learning rate that satisfies the strong Wolfe conditions. This method maintains a positive definite approximation of the inverse Hessian matrix."backtracking"— Search for a learning rate that satisfies sufficient decrease conditions. This method does not maintain a positive definite approximation of the inverse Hessian matrix.
You can specify Line search method only when Training
algorithm is LBFGS.
Number of state updates to store, specified as a positive integer. Values between 3 and 20 suit most tasks.
The L-BFGS algorithm uses a history of gradient calculations to approximate the Hessian matrix recursively. For more information, see Limited-Memory BFGS (Deep Learning Toolbox).
You can specify History size only when Training
algorithm is LBFGS.
Initial value that characterizes the approximate inverse Hessian matrix, specified as a positive scalar.
To save memory, the L-BFGS algorithm does not store and invert the dense Hessian matrix B. Instead, the algorithm uses the approximation , where m is the history size, the inverse Hessian factor is a scalar, and I is the identity matrix. The algorithm then stores the scalar inverse Hessian factor only. The algorithm updates the inverse Hessian factor at each step.
The initial inverse hessian factor is the value of .
For more information, see Limited-Memory BFGS (Deep Learning Toolbox).
You can specify Initial inverse Hessian factor only when
Training algorithm is LBFGS.
Maximum number of line search iterations to determine the learning rate, specified as a positive integer.
You can specify Maximum line search iterations only when
Training algorithm is LBFGS.
Learning rate used for training, specified as a positive scalar. The default value
is 0.001 for Adam and RMSProp solvers and 0.01 for
SGDM solver.
If the learning rate is too small, then training can take a long time. If the
learning rate is too large, then training might reach a suboptimal result or diverge.
You can specify Learn rate only when Training
algorithm is ADAM,
SGDM, or RMSProp.
Maximum number of epochs to use for training, specified as a positive integer. An
epoch is the full pass of the training algorithm over the entire training set. You can
specify Maximum number of epochs only when Training
algorithm is ADAM,
SGDM, or RMSProp.
Size of the mini-batch to use for each training iteration, specified as a positive integer. A mini-batch is a subset of the training set that is used to evaluate the gradient of the loss function and update the weights.
If the mini-batch size does not evenly divide the number of training samples, then
the estimation process discards the training data that does not fit into the final
complete mini-batch of each epoch. You can specify Size of
mini-batch only when Training algorithm is
ADAM, SGDM, or
RMSProp.
Number of samples in each frame or batch when segmenting data for model training, specified as a positive integer.
Number of samples in the overlap between successive frames when segmenting data for model training, specified as an integer. A negative integer indicates that certain data samples are skipped when creating the data frames.
You can select one of the following options:
zoh— Zero-order hold interpolation methodfoh— First-order hold interpolation methodcubic— Cubic interpolation methodmakima— Modified Akima interpolation methodpchip— Shape-preserving piecewise cubic interpolation methodspline— Spline interpolation method (default)
This is the interpolation method used to interpolate the input when integrating
continuous-time neural state-space models. For more information, see interpolation
methods in interp1.
Enable displaying a validation plot periodically during training. The validation plot shows a comparison between the predicted output response to measured validation inputs and the measured validation outputs. The plot also displays the model fit percentage.
This is the number of epochs after which the validation plot is updated with a new
comparison (new predicted output against measured outputs). For example, if
Validation data fit frequency is 10, the validation plot is
updated every 10 epochs. For more information, see nlssest.
Enable displaying a training plot during training (estimation). The training plot shows how the state and output network loss values evolve after each training epoch.
Display results
After estimation (training), plot a comparison between the predicted output response to measured estimation inputs and the measured estimation outputs. Selecting this parameter also displays the model fit percentage.
After estimation (training), plot a comparison between the predicted output response to measured validation inputs and the measured validation outputs. Selecting this parameter also displays the model fit percentage. This parameter is available only if you select validation data in the Select Data section.
Version History
Introduced in R2023bThe Estimate Neural State-Space Model Live Editor task now supports multi-experiment data.
See Also
Objects
idNeuralStateSpace|nssTrainingADAM|nssTrainingSGDM|nssTrainingRMSProp|nssTrainingLBFGS|idss|idnlgrey
Functions
createMLPNetwork|setNetwork|nssTrainingOptions|nlssest|generateMATLABFunction|idNeuralStateSpace/evaluate|idNeuralStateSpace/linearize|sim
Blocks
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)