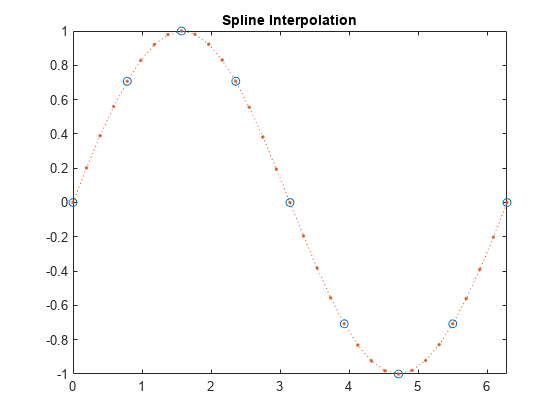
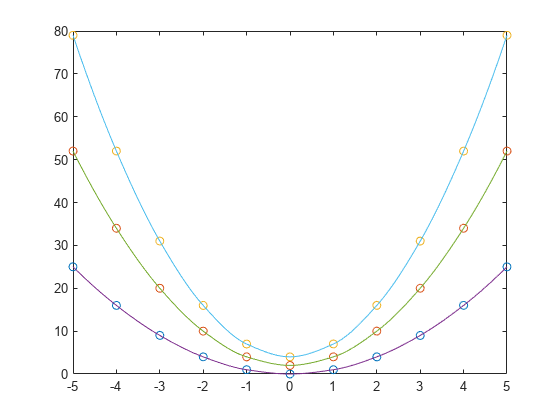
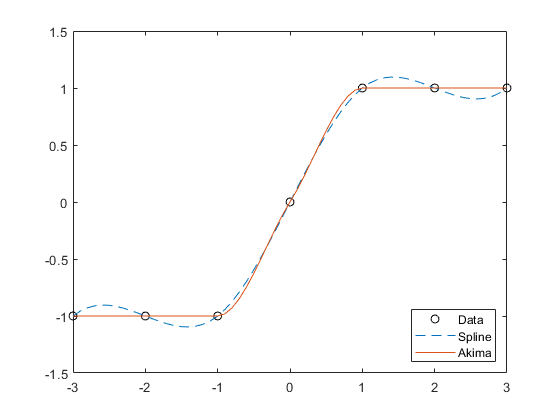
interp1
1-D data interpolation (table lookup)
Syntax
Description
vq = interp1(x,v,xq)interp1 uses linear interpolation. Vector
x contains the sample points, and v
contains the corresponding values, v(x).
Vector xq contains the coordinates of the query
points.
If you have multiple sets of data that are sampled at the same point
coordinates, then you can pass v as an array. Each column of
array v contains a different set of 1-D sample values.
vq = interp1(x,v,xq,method,extrapolation)x. Set extrapolation to
'extrap' when you want to use the
method algorithm for extrapolation. Alternatively, you
can specify a scalar value, in which case, interp1 returns
that value for all points outside the domain of x.
vq = interp1(v,xq)1 to n, where n
depends on the shape of v:
When v is a vector, the default points are
1:length(v).When v is an array, the default points are
1:size(v,1).
Use this syntax when you are not concerned about the absolute distances between points.
vq = interp1(v,xq,method,extrapolation)
pp = interp1(x,v,method,'pp')method algorithm.
Note
This syntax is not recommended. Use griddedInterpolant
instead.
Examples
Input Arguments
Output Arguments
More About
References
[1] Akima, Hiroshi. "A new method of interpolation and smooth curve fitting based on local procedures." Journal of the ACM (JACM) , 17.4, 1970, pp. 589-602.
[2] Akima, Hiroshi. "A method of bivariate interpolation and smooth surface fitting based on local procedures." Communications of the ACM , 17.1, 1974, pp. 18-20.