deepSignalAnomalyDetector
Syntax
Description
d = deepSignalAnomalyDetector(numChannels)d based on a 1-D convolutional
autoencoder.
d = deepSignalAnomalyDetector(numChannels,modelType)
d = deepSignalAnomalyDetector(___,Name=Value)
Examples
Load the file sineWaveAnomalyData.mat, which contains two sets of synthetic three-channel sinusoidal signals.
sineWaveNormal contains 10 sinusoids of stable frequency and amplitude. Each signal has a series of small-amplitude impact-like imperfections. The signals have different lengths and initial phases. Plot the first three normal signals.
load sineWaveAnomalyData.mat sl = 3; tiledlayout("vertical") ax = zeros(sl,1); for kj = 1:sl ax(kj) = nexttile; plot(sineWaveNormal{kj}) title("Normal Signal") end

sineWaveAbnormal contains three signals, all of the same length. Each signal in the set has one or more anomalies.
All channels of the first signal have an abrupt change in frequency that lasts for a finite time.
The second signal has a finite-duration amplitude change in one of its channels.
The third signal has spikes at random times in all channels.
Plot the three signals with anomalies.
for kj = 1:sl plot(ax(kj),sineWaveAbnormal{kj}) title(ax(kj),"Signal with Anomalies") end

Create an autoencoder object to detect the anomalies in the abnormal signals. The object contains a neural network that you can train to best reproduce an input set of anomaly-free data. The trained object encodes the patterns inherent in the data and can recognize if a signal contains regions that deviate from the patterns. You do not need to label any data to train the detector. The training process is unsupervised.
By default, the deepSignalAnomalyDetector function creates objects with convolutional autoencoders. The only mandatory argument is the number of channels in the training and testing signals.
D = deepSignalAnomalyDetector(sl)
D =
deepSignalAnomalyDetectorCNN with properties:
IsTrained: 0
NumChannels: 3
Model Information
ModelType: 'convautoencoder'
FilterSize: 8
NumFilters: 32
NumDownsampleLayers: 2
DownsampleFactor: 2
DropoutProbability: 0.2000
Threshold Information
Threshold: []
ThresholdMethod: 'contaminationFraction'
ThresholdParameter: 0.0100
Window Information
WindowLength: 1
OverlapLength: 'auto'
WindowLossAggregation: 'mean'
Use the trainDetector function to train the convolutional neural network with the normal data. Use the training options for the adaptive moment estimation (Adam) optimizer. Specify a maximum number of 100 epochs to use for training. For more information, see trainingOptions (Deep Learning Toolbox).
opts = trainingOptions("adam",MaxEpochs=100);
trainDetector(D,sineWaveNormal,opts) Iteration Epoch TimeElapsed LearnRate TrainingLoss
_________ _____ ___________ _________ ____________
1 1 00:00:01 0.001 0.49381
50 50 00:00:05 0.001 0.06591
100 100 00:00:07 0.001 0.047143
Training stopped: Max epochs completed
Computing threshold...
Threshold computation completed.
Use the trained detector to detect the anomalies in the abnormal data. The detect function outputs two cell arrays, with each array element corresponding to a signal in the testing data.
[lbls,loss] = detect(D,sineWaveAbnormal);
The first output of detect is a categorical array that declares each sample of a signal as being anomalous or not. A sample can be either a point, a signal region, or an entire signal. An anomaly is detected when the reconstruction loss, or the difference between the value of a signal and the value reconstructed by the detector based on the training data, exceeds a given threshold. You can set a threshold manually or you can direct the detector to compute a threshold based on a specified statistic of loss values computed on training data.
for kj = 1:sl hold(ax(kj),"on") plot(ax(kj),lbls{kj},LineWidth=2) title(ax(kj),"Signal with Anomalies") hold(ax(kj),"off") end

Alternatively, use the plotAnomalies function to plot each channel of each signal and the anomalies found by the detector. The detector decides that there is an anomaly in a signal if it detects one in any of the signal channels. plotAnomalies plots the anomaly locations in all channels.
for kj = 1:sl figure plotAnomalies(D,sineWaveAbnormal{kj}) end



The second output of detect is the computed reconstruction loss that the object uses to determine the labeling. You can plot the reconstruction loss directly from the output of detect, but it is more convenient to use the plotLoss function, which also displays the threshold value above which a sample is declared to be anomalous.
tiledlayout("vertical") for kj = 1:sl nexttile plotLoss(D,sineWaveAbnormal{kj}) end

The plotLossDistribution function displays the reconstruction loss distribution and gives insight into the way the detector chooses the threshold. You can plot the distribution loss for the normal data alongside the distribution loss for the abnormal data. The threshold chosen by the detector is larger than the loss values for most of the normal-data samples but smaller than an appreciable number of abnormal-data losses that signal the possible presence of anomalies. You can improve the choice of threshold by using the updateDetector function.
clf plotLossDistribution(D,sineWaveNormal,sineWaveAbnormal)

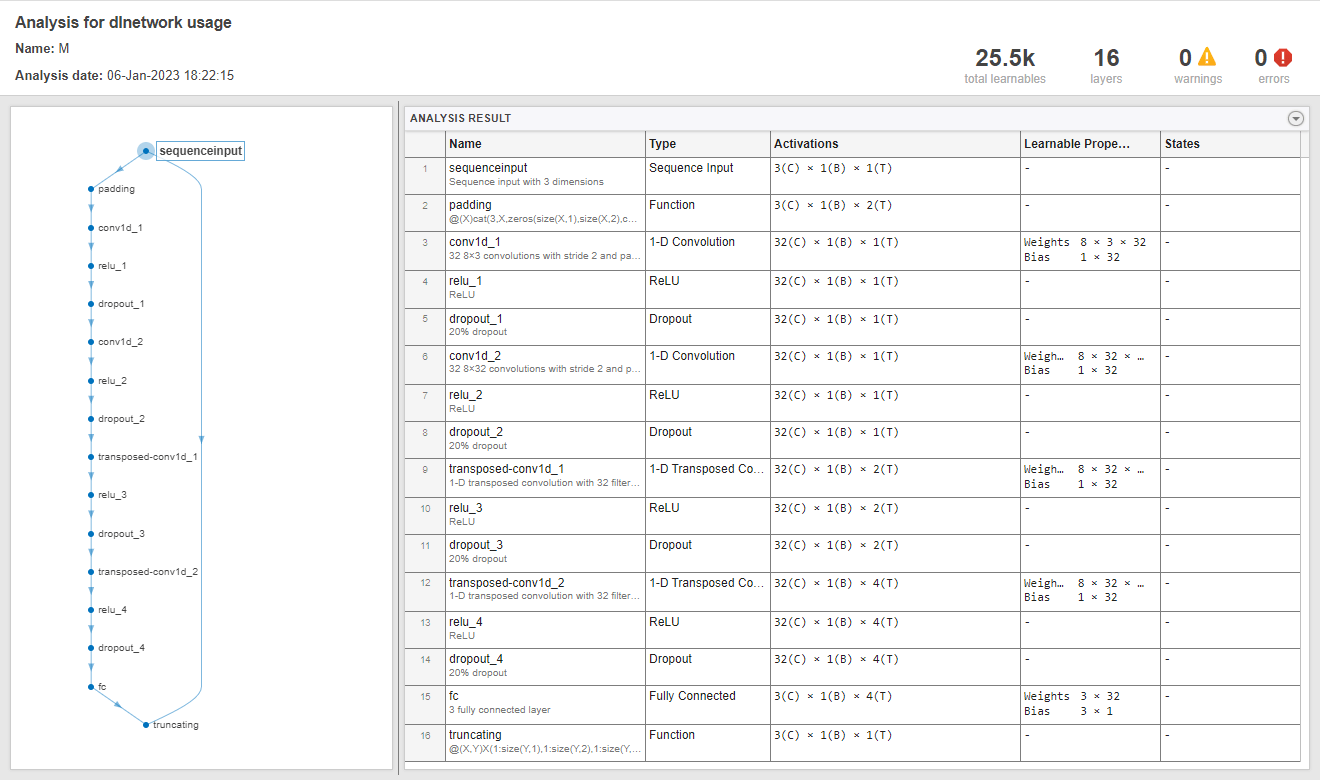
Use the getModel function to obtain the underlying neural network model corresponding to the detector. You can then inspect the model using analyzeNetwork (Deep Learning Toolbox).
M = getModel(D); analyzeNetwork(M)
Input Arguments
Number of channels in each signal input to the detector d,
specified as a positive integer.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Type of deep learning model implemented by the detector d,
specified as one of these:
"convautoencoder"— Implement a 1-D convolutional autoencoder."lstmautoencoder"— Implement a long short-term memory (LSTM) autoencoder."lstmforecaster"— Implement a long short-term memory (LSTM) forecaster.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: ThresholdMethod="manual",Threshold=0.4 specifies that the
threshold value above which a sample is declared to be anomalous is set manually to
0.4.
Window
Window length of each signal segment, specified as a positive integer or as "fullSignal".
If you specify
WindowLengthas an integer, the detector divides each input signal into segments. The length of each segment is equal to the specified value in samples.If you specify
WindowLengthas"fullSignal", the detector treats each input signal as a single segment.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | char | string
Number of overlapped samples between window segments, specified as a positive integer or as "auto".
If you specify
WindowLengthandOverlapLengthas integers, the detector sets the number of overlapped samples to the specified value. The number of overlapped samples must be less than the window length.If you specify
WindowLengthas an integer andOverlapLengthas"auto", the detector sets the number of overlapped samples toWindowLength– 1.If you specify
WindowLengthas"fullSignal", you cannot specifyOverlapLengthas an integer.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | logical | char | string
Method to aggregate sample loss within each window segment, specified as one of these:
"max"— Compute the aggregated window loss as the maximum value of all the sample losses within the window."mean"— Compute the aggregated window loss as the mean value of all the sample losses within the window."median"— Compute the aggregated window loss as the median value of all the sample losses within the window."min"— Compute the aggregated window loss as the minimum value of all the sample losses within the window.
The detector computes the detection loss of each sample within a window segment and aggregates the loss values over each window.
Threshold
Method to compute the detection threshold, specified as one of these:
"contaminationFraction"— Value corresponding to the detection of anomalies within a specified fraction of windows. The fraction value is specified byThresholdParameter."max"— Maximum window loss measured over the entire training data set and multiplied byThresholdParameter."median"— Median window loss measured over the entire training data set and multiplied byThresholdParameter."mean"— Mean window loss measured over the entire training data set and multiplied byThresholdParameter."manual"— Manual detection threshold value based onThreshold."customFunction"— Custom detection threshold value based onThresholdFunction.
If you specify ThresholdMethod, you can also specify ThresholdParameter, Threshold, or ThresholdFunction. The available threshold parameter depends on the specified detection method.
Detection threshold, specified as a real scalar.
If
ThresholdMethodis specified as"max","mean", or"median", specifyThresholdParameteras a positive scalar. If you do not specifyThresholdParameter, the detector sets the threshold to 1.If
ThresholdMethodis specified as"contaminationFraction", specifyThresholdParameteras a nonnegative scalar less than 0.5. If you do not specifyThresholdParameter, the detector sets the threshold to 0.01.If
ThresholdMethodis specified as"customFunction"or"manual", this argument does not apply.
Manual detection threshold, specified as a positive scalar. This argument applies only when ThresholdMethod is specified as "manual".
Use this option when you do not want the detector to compute a threshold based on training data.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Function to compute custom detection threshold, specified as a function handle. This argument applies only when ThresholdMethod is specified as "customFunction".
The function must have two inputs:
The first input is a cell array of aggregated window loss values.
The second input is a cell array of sample loss values before aggregation.
Each cell contains a loss vector for one signal observation.
The function must return a positive scalar corresponding to the detection threshold.
Use this option when you want to compute a threshold based on training data.
Data Types: function_handle
Convolutional Encoder
Number of convolutional layers in the downsampling section of the model, specified as a positive integer.
The model uses the same number of transposed convolutional layers in the upsampling section, with the parameters in reverse order.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Downsampling factor used by convolutional layers, specified as a positive integer or a vector of positive integers.
If you specify
DownsampleFactoras a scalar, each layer uses the same downsampling factor.If you specify
DownsampleFactoras a vector, the ith layer uses a downsampling factor equal to the value of the ith vector element. The length of the vector must be equal to the number of layers specified byNumDownsampleLayers.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Number of filters in convolutional layers, specified as a positive scalar integer or a vector of positive integers.
If you specify
NumFiltersas a scalar, each layer has the same number of filters.If you specify
NumFiltersas a vector, the ith layer has a number of filters equal to the value of the ith vector element. The length of the vector must be equal to the number of layers specified byNumDownsampleLayers.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Size of filters in convolutional layers, specified as a positive integer or a vector of positive integers.
If you specify
FilterSizeas a scalar, the size of each filter is the same in all layers.If you specify
FilterSizeas a vector, the size of the filters in the ith layer is equal to the value of the ith vector element. The length of the vector must be equal to the number of layers specified byNumDownsampleLayers.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Dropout probability used to avoid overfitting, specified as a nonnegative scalar less than 1.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
LSTM Autoencoder
Number of hidden units for each LSTM layer in the encoder, specified as a vector of positive integers. The ith element of the vector sets the number of hidden units in the ith layer.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Number of hidden units in each LSTM layer of the decoder, specified as a vector of positive integers. The ith element of the vector sets the number of hidden units in the ith layer.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
LSTM Forecaster
Since R2024a
This property is read-only.
Number of steps ahead for forecasting, specified as a positive integer scalar.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
This property is read-only.
Number of hidden units of LSTM layers, specified as a positive integer scalar or a vector of positive integers. The ith element of the vector sets the number of hidden units in the ith layer.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Output Arguments
Signal anomaly detector, returned as a deepSignalAnomalyDetectorCNN object, a deepSignalAnomalyDetectorLSTM object, or a deepSignalAnomalyDetectorLSTMForecaster object. You must train
d with data before you can use it to detect anomalies.
Algorithms
To detect anomalies, deepSignalAnomalyDetector divides each channel
of each signal into possibly overlapping windows. You can specify the window length using
WindowLength and the number of overlap samples using
OverlapLength.
By default, the detector uses signals with a length of one sample and zero samples of overlap between adjoining windows. These settings detect point anomalies.
You can also set
WindowLengthto"fullSignal"to detect whole-signal anomalies.
For each window, deepSignalAnomalyDetector computes
sample-by-sample losses, defined as the square of the difference in
value between the original signal and the reconstructed signal. The detector then aggregates
the losses using the method specified using WindowLossAggregation. You
can use the maximum sample loss, the minimum sample loss, the mean of the losses, or the
median of the losses.
If the aggregated loss for a window is larger than a specified threshold, the object detects the whole window as abnormal.
To determine the threshold, deepSignalAnomalyDetector finds the
aggregated losses for all the windows in the training data and combines them into a
statistic. You can specify the method that the detector uses to compute the threshold using
ThresholdMethod.
The statistic can be the maximum value of the losses across the data, the minimum value, the mean value, or the median value. The threshold is the statistic multiplied by
ThresholdParameter.If you expect a given fraction of windows in your data to be abnormal, you can instead specify
ThresholdMethodascontaminationFractionandThresholdParameteras the expected fraction. In that case, the object sets the threshold to the loss value such that the number of windows with greater losses is equal to the specified fraction of the total.If you prefer, you can specify a threshold manually by specifying
ThresholdMethodas"manual"and using theThresholdargument. You can also use your own custom function to compute the threshold.
When the trained object detects a window as normal or abnormal, you can specify whether
to associate the label with the whole window or with each of its samples. If you choose to
divide your signals into overlapping windows, a conflict can arise because some signal
samples can at the same time be in a window detected as normal and in a window detected as
abnormal. In that case, you can use the OverlapPriority
argument of the detect function
to declare samples with conflicting labels as either normal or abnormal.
Extended Capabilities
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2023aUsing trainingOptions with an integer
SequenceLength value produces fewer batches than in previous releases
and the training results can be different. To achieve a similar number of iterations as
before, increase the number of epochs.
deepSignalAnomalyDetector has a third mode,
deepSignalAnomalyDetectorLSTMForecaster, that uses a long short-term
memory network to detect anomalies in streaming workflows and thus in real time.
The saveModel
object function creates a MAT file containing the trained network and parameters
corresponding to any object generated by deepSignalAnomalyDetector. Use the generated
file with the Deep Signal Anomaly Detector
Simulink® block.
See Also
Objects
deepSignalAnomalyDetectorCNN|deepSignalAnomalyDetectorLSTM|deepSignalAnomalyDetectorLSTMForecaster
Functions
Blocks
- Deep Signal Anomaly Detector (DSP System Toolbox)
Topics
- Detect Anomalies in Signals Using deepSignalAnomalyDetector
- Detect Anomalies in Machinery Using LSTM Autoencoder
- Detect Anomalies in ECG Data Using Wavelet Scattering and LSTM Autoencoder in Simulink (DSP System Toolbox)
- Detect Anomalies in Industrial Machinery Using Three-Axis Vibration Data (Predictive Maintenance Toolbox)
- Anomaly Detection Using Convolutional Autoencoder with Wavelet Scattering Sequences
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)