Create Training Data for 3-D Medical Image Semantic Segmentation
This example shows how to create training data for a 3-D semantic segmentation network using a groundTruthMedical object that contains 3-D medical image data. This example also shows how to transform datastores to preprocess and augment image and pixel label data.
Create Ground Truth Object
You can export a groundTruthMedical object from the Medical Image Labeler app or create one programmatically.
This example creates a groundTruthMedical object by using the createGroundTruthMed3D helper function. The helper function is attached to this example as a supporting file. The ground truth object references three data sources, including one chest CT volume stored as a directory of DICOM files and two chest CT volumes stored as NIfTI files. The directory of DICOM files and its label image are attached to this example as supporting files. The createGroundTruthMed3D function downloads the NIfTI files and their label images from the MathWorks® website. The NIfTI files are a subset of the Medical Segmentation Decathlon data set [1], and have a total file size of approximately 76 MB.
gTruthMed = createGroundTruthMed3D;
Extract Data from Ground Truth Object
Extract the data source and label image file names from the groundTruthMedical object.
dataSource = gTruthMed.DataSource.Source; labelData = gTruthMed.LabelData;
Remove data sources that are missing label images.
noLabelsIdx = labelData~="";
dataSource = dataSource(noLabelsIdx);
labelData = labelData(noLabelsIdx);Extract the label definitions from the groundTruthMedical object. Add an addition label definition for the background region, corresponding to a pixel value of 0.
labelDefs = gTruthMed.LabelDefinitions;
labelDefs(2,:) = {"background",[0 1 0],0};Load Data Source Images into Image Datastore
A groundTruthMedical object can contain data sources stored as single DICOM, NIFTI, or NRRD file, or a directory of DICOM files. The imageDatastore object does not support reading volumetric images from a directory of DICOM files. Use the convertMultifileDICOMs supporting function to search dataSource for data sources stored as a directory of DICOM files, and convert any DICOM directories into single MAT files.
dataSource = convertMultifileDICOMs(dataSource); dataSource = string(dataSource);
Load the updated data sources into an imageDatastore. Specify a custom read function, readMedicalVolumes, which is defined at the end of this example. The readMedicalVolumes function reads data from medical volumes stored as a single MAT file, DICOM file, or NIfTI file. The .gz file extension corresponds to compressed NIfTI files.
imds = imageDatastore(dataSource,... ReadFcn=@readMedicalVolumes,... FileExtensions=[".mat",".dcm",".nii",".gz"]);
Load Label Images into Pixel Label Datastore
Load the label images into a pixelLabelDatastore (Computer Vision Toolbox) using niftiread as the read function.
pxds = pixelLabelDatastore(cellstr(labelData),labelDefs.Name,labelDefs.PixelLabelID,... ReadFcn=@niftiread,... FileExtensions=[".nii",".gz"]);
Preview one image volume and its label image.
vol = preview(imds); label = preview(pxds);
Display one slice of the previewed volume by using the labeloverlay function. To better view the grayvalues of the CT slice with labeloverlay, first rescale the image intensities to the range [0, 1].
volslice = vol(:,:,100); volslice = rescale(volslice); labelslice = label(:,:,100); imOverlay = labeloverlay(volslice,labelslice); imshow(imOverlay)
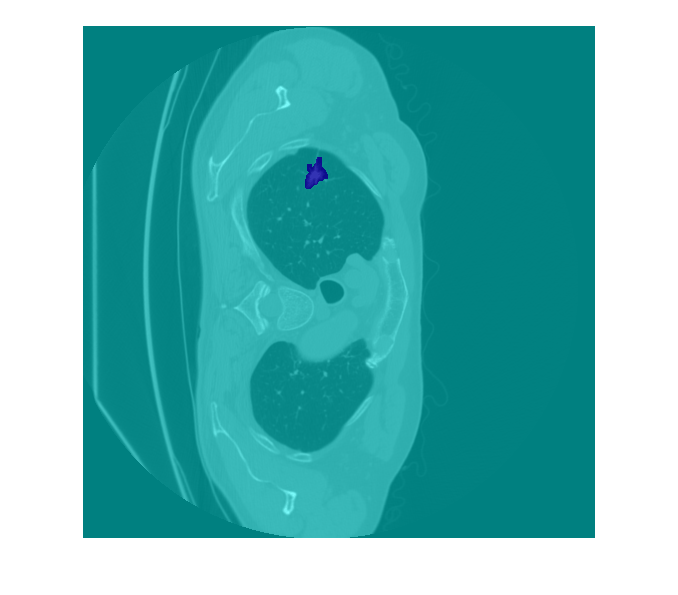
Pair Image and Label Data
Create a CombinedDatastore that pairs each data source image with its corresponding label image.
trainingData = combine(imds,pxds);
Augment and Preprocess Training Data
Specify the input size of the target deep learning network.
targetSize = [300 300 60];
Augment the training data by using the transform function with custom operations specified by the jitterImageIntensityAndWarp supporting function defined a the end of this example.
augmentedTrainingData = transform(trainingData,@jitterImageIntensityAndWarp);
Preprocess the training data by using the transform function with custom preprocessing operation specified by the centerCropImageAndLabel supporting function defined at the end of this example.
preprocessedTrainingData = transform(augmentedTrainingData,...
@(data)centerCropImageAndLabel(data,targetSize));Supporting Functions
The convertMultifileDICOMs function reads a cell array of data source file names, and converts any data sources stored as a directory of DICOM files into a single MAT file.
function dataSource = convertMultifileDICOMs(dataSource) numEntries = length(dataSource); dataFileDir = fullfile(pwd,"GroundTruthData"); if ~isfolder(dataFileDir) mkdir(dataFileDir) end for idx = 1:numEntries currEntry = dataSource{idx}; % Multi-file DICOMs if length(currEntry) > 1 matFileName = fileparts(currEntry(1)); matFileName = split(matFileName,filesep); matFileName = replace(strtrim(matFileName(end))," ","_"); matFileName = strcat(fullfile(dataFileDir,matFileName),".mat"); vol = medicalVolume(currEntry); data = vol.Voxels; save(matFileName,"data"); dataSource{idx} = string(matFileName); end end end
The readMedicalVolumes function loads medical image volume data from a single MAT file, DICOM file, or NIfTI file.
function data = readMedicalVolumes(filename) [~,~,ext] = fileparts(filename); if ext == ".mat" d = load(filename); data = d.data; else vol = medicalVolume(filename); data = vol.Voxels; end end
The jitterImageColorAndWarp function randomly adjusts the brightness, contrast, and gamma correction of the data source image values, and applies random geometric transformations including scaling, reflection, and rotation to the data source and pixel label images.
function out = jitterImageIntensityAndWarp(data) % Unpack original data. I = data{1}; C = data{2}; % Apply random intensity jitter. I = jitterIntensity(I,Brightness=0.3,Contrast=0.4,Gamma=0.2); % Define random affine transform. tform = randomAffine3d(Scale=[0.8 1.5],XReflection=true,Rotation=[-30 30]); rout = affineOutputView(size(I),tform); % Transform image and pixel labels. augmentedImage = imwarp(I,tform,"OutputView",rout); augmentedLabel = imwarp(C,tform,"OutputView",rout); % Return augmented data. out = {augmentedImage,augmentedLabel}; end
The centerCropImageAndLabel function crops the data source images and pixel labels to the input size of the target deep learning network.
function out = centerCropImageAndLabel(data,targetSize) win = centerCropWindow3d(size(data{1}),targetSize); out{1} = imcrop3(data{1},win); out{2} = imcrop3(data{2},win); end
References
[1] Medical Segmentation Decathlon. "Brain Tumours." Tasks. Accessed May 10, 2018. http://medicaldecathlon.com/.
The Medical Segmentation Decathlon data set is provided under the CC-BY-SA 4.0 license. All warranties and representations are disclaimed. See the license for details.
See Also
groundTruthMedical | imageDatastore | pixelLabelDatastore (Computer Vision Toolbox) | transform | combine