featureInputLayer
Feature input layer
Description
A feature input layer inputs feature data to a neural network and applies data normalization. Use this layer when you have a data set of numeric scalars representing features (data without spatial or time dimensions).
For image input, use imageInputLayer.
Creation
Description
layer = featureInputLayer(numFeatures)InputSize property to the specified number of features.
layer = featureInputLayer(numFeatures,Name=Value)
Input Arguments
Number of features for each observation in the data, specified as a positive integer.
For image input, use imageInputLayer.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and
enclose
Name
in quotes.
Example: featureInputLayer(21,Name="input") creates a feature input
layer with number of features set to 21 and name
'input'.
Data normalization to apply every time data is forward propagated through the input layer, specified as one of the following:
"zerocenter"— Subtract the mean specified byMean."zscore"— Subtract the mean specified byMeanand divide byStandardDeviation."rescale-symmetric"— Rescale the input to be in the range [-1, 1] using the minimum and maximum values specified byMinandMax, respectively."rescale-zero-one"— Rescale the input to be in the range [0, 1] using the minimum and maximum values specified byMinandMax, respectively."none"— Do not normalize the input data.function handle — Normalize the data using the specified function. The function must be of the form
Y = f(X), whereXis the input data and the outputYis the normalized data.
If the input data is complex-valued
and the SplitComplexInputs option is 0
(false), then the Normalization option
must be "zerocenter", "zscore",
"none", or a function handle. (since R2024a)
Before R2024a: To input complex-valued data into the
network, the SplitComplexInputs option must be
1 (true).
Tip
The software, by default, automatically calculates the normalization
statistics when you use the trainnet function. To save time when training, specify the
required statistics for normalization and set the ResetInputNormalization option in trainingOptions to 0
(false).
The FeatureInputLayer object stores the
Normalization property as a character vector or a function
handle.
Normalization dimension, specified as one of the following:
"auto"– If theResetInputNormalizationtraining option is0(false) and you specify any of the normalization statistics (Mean,StandardDeviation,Min, orMax), then normalize over the dimensions matching the statistics. Otherwise, recalculate the statistics at training time and apply channel-wise normalization."channel"– Channel-wise normalization."all"– Normalize all values using scalar statistics.
The FeatureInputLayer object stores this property as a character
vector.
Mean for zero-center and z-score normalization, specified as a 1-by-numFeatures vector of means per feature, a numeric scalar, or [].
To specify the Mean property, the Normalization property must be "zerocenter"
or "zscore". If Mean is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the mean using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to0.
Mean can be complex-valued. (since R2024a) If
Mean is complex-valued, then the
SplitComplexInputs option must be 0
(false).
Before R2024a: Split the mean into real and imaginary parts and split
the input data into real and imaginary parts by setting the
SplitComplexInputs option to
1 (true).
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Complex Number Support: Yes
Standard deviation for z-score normalization, specified as a 1-by-numFeatures vector of means per feature, a numeric scalar, or [].
To specify the StandardDeviation property, the
Normalization property must be
"zscore". If StandardDeviation is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the standard deviation using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to1.
StandardDeviation can be
complex-valued. (since R2024a) If StandardDeviation is complex-valued, then
the SplitComplexInputs option must be 0
(false).
Before R2024a: Split the standard deviation into real and imaginary
parts and split the input data into real and imaginary parts by setting the
SplitComplexInputs option to 1
(true).
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Complex Number Support: Yes
Minimum value for rescaling, specified as a 1-by-numFeatures vector of minima per feature, a numeric scalar, or [].
To specify the Min property, the Normalization must be "rescale-symmetric" or
"rescale-zero-one". If Min is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the minimum value using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to-1and0whenNormalizationis"rescale-symmetric"and"rescale-zero-one", respectively.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Maximum value for rescaling, specified as a 1-by-numFeatures vector of maxima per feature, a numeric scalar, or [].
To specify the Max property, the Normalization must be "rescale-symmetric" or
"rescale-zero-one". If Max is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the maximum value using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to1.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Flag to split input data into real and imaginary components specified as one of these values:
0(false) – Do not split input data.1(true) – Split data into real and imaginary components.
When SplitComplexInputs is 1, then the
layer outputs twice as many channels as the input data. For example, if the input
data is complex-valued with numChannels channels, then the layer
outputs data with 2*numChannels channels, where channels
1 through numChannels contain the real
components of the input data and numChannels+1 through
2*numChannels contain the imaginary components of the input
data. If the input data is real, then channels numChannels+1
through 2*numChannels are all zero.
If the input data is complex-valued and
SplitComplexInputs is 0
(false), then the layer passes the complex-valued data to the
next layers. (since R2024a)
Before R2024a: To input complex-valued data into a
neural network, the SplitComplexInputs option of the input
layer must be 1 (true).
For an example showing how to train a network with complex-valued data, see Train Network with Complex-Valued Data.
Properties
Feature Input
Number of features for each observation in the data, specified as a positive integer.
For image input, use imageInputLayer.
Data normalization to apply every time data is forward propagated through the input layer, specified as one of the following:
"zerocenter"— Subtract the mean specified byMean."zscore"— Subtract the mean specified byMeanand divide byStandardDeviation."rescale-symmetric"— Rescale the input to be in the range [-1, 1] using the minimum and maximum values specified byMinandMax, respectively."rescale-zero-one"— Rescale the input to be in the range [0, 1] using the minimum and maximum values specified byMinandMax, respectively."none"— Do not normalize the input data.function handle — Normalize the data using the specified function. The function must be of the form
Y = f(X), whereXis the input data and the outputYis the normalized data.
If the input data is complex-valued and the
SplitComplexInputs option is 0
(false), then the Normalization option must be
"zerocenter", "zscore",
"none", or a function handle. (since R2024a)
Before R2024a: To input complex-valued data into the network,
the SplitComplexInputs option must be 1
(true).
Tip
The software, by default, automatically calculates the normalization statistics when you use
the trainnet
function. To save time when training, specify the required statistics for normalization
and set the ResetInputNormalization option in trainingOptions to 0
(false).
The FeatureInputLayer object stores this property as a character vector or a
function handle.
Normalization dimension, specified as one of the following:
"auto"– If theResetInputNormalizationtraining option is0(false) and you specify any of the normalization statistics (Mean,StandardDeviation,Min, orMax), then normalize over the dimensions matching the statistics. Otherwise, recalculate the statistics at training time and apply channel-wise normalization."channel"– Channel-wise normalization."all"– Normalize all values using scalar statistics.
The FeatureInputLayer object stores this property as a character vector.
Mean for zero-center and z-score normalization, specified as a 1-by-numFeatures vector of means per feature, a numeric scalar, or [].
To specify the Mean property, the Normalization property must be "zerocenter"
or "zscore". If Mean is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the mean using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to0.
Mean can be complex-valued. (since R2024a) If
Mean is complex-valued, then the
SplitComplexInputs option must be 0
(false).
Before R2024a: Split the mean into real and imaginary parts and split
the input data into real and imaginary parts by setting the
SplitComplexInputs option to
1 (true).
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Complex Number Support: Yes
Standard deviation for z-score normalization, specified as a 1-by-numFeatures vector of means per feature, a numeric scalar, or [].
To specify the StandardDeviation property, the
Normalization property must be
"zscore". If StandardDeviation is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the standard deviation using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to1.
StandardDeviation can be
complex-valued. (since R2024a) If StandardDeviation is complex-valued, then
the SplitComplexInputs option must be 0
(false).
Before R2024a: Split the standard deviation into real and imaginary
parts and split the input data into real and imaginary parts by setting the
SplitComplexInputs option to 1
(true).
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Complex Number Support: Yes
Minimum value for rescaling, specified as a 1-by-numFeatures vector of minima per feature, a numeric scalar, or [].
To specify the Min property, the Normalization must be "rescale-symmetric" or
"rescale-zero-one". If Min is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the minimum value using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to-1and0whenNormalizationis"rescale-symmetric"and"rescale-zero-one", respectively.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Maximum value for rescaling, specified as a 1-by-numFeatures vector of maxima per feature, a numeric scalar, or [].
To specify the Max property, the Normalization must be "rescale-symmetric" or
"rescale-zero-one". If Max is
[], then the software automatically sets the property at training or
initialization time:
The
trainnetfunction calculates the maximum value using the training data and uses the resulting value.The
initializefunction and thedlnetworkfunction when theInitializeoption is1(true) sets the property to1.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
This property is read-only.
Flag to split input data into real and imaginary components specified as one of these values:
0(false) – Do not split input data.1(true) – Split data into real and imaginary components.
When SplitComplexInputs is 1, then the layer
outputs twice as many channels as the input data. For example, if the input data is
complex-valued with numChannels channels, then the layer outputs data
with 2*numChannels channels, where channels 1
through numChannels contain the real components of the input data and
numChannels+1 through 2*numChannels contain
the imaginary components of the input data. If the input data is real, then channels
numChannels+1 through 2*numChannels are all
zero.
If the input data is complex-valued and
SplitComplexInputs is 0
(false), then the layer passes the complex-valued data to the
next layers. (since R2024a)
Before R2024a: To input complex-valued data into a neural
network, the SplitComplexInputs option of the input layer must be
1 (true).
For an example showing how to train a network with complex-valued data, see Train Network with Complex-Valued Data.
Layer
This property is read-only.
Number of inputs of the layer. The layer has no inputs.
Data Types: double
This property is read-only.
Input names of the layer. The layer has no inputs.
Data Types: cell
This property is read-only.
Number of outputs from the layer, stored as 1. This layer has a
single output only.
Data Types: double
This property is read-only.
Output names, stored as {'out'}. This layer has a single output
only.
Data Types: cell
Examples
Create a feature input layer with the name "input" for observations consisting of 21 features.
layer = featureInputLayer(21,Name="input")layer =
FeatureInputLayer with properties:
Name: 'input'
InputSize: 21
SplitComplexInputs: 0
Hyperparameters
Normalization: 'none'
NormalizationDimension: 'auto'
Include a feature input layer in a Layer array.
numFeatures = 21;
numClasses = 3;
layers = [
featureInputLayer(numFeatures)
fullyConnectedLayer(numClasses)
softmaxLayer]layers =
3×1 Layer array with layers:
1 '' Feature Input 21 features
2 '' Fully Connected 3 fully connected layer
3 '' Softmax softmax
Define the size of the input image, the number of features of each observation, the number of classes, and the size and number of filters of the convolution layer.
imageInputSize = [28 28 1]; numFeatures = 1; numClasses = 10; filterSize = 5; numFilters = 16;
To create a network with two inputs, define the network in two parts and join them, for example, by using a concatenation layer.
Create a dlnetwork object.
net = dlnetwork;
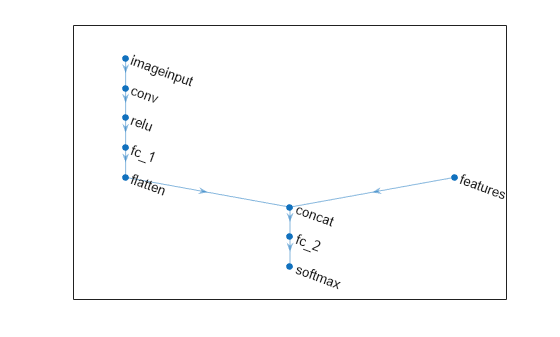
Define the first part of the network. Define the image classification layers and include a flatten layer and a concatenation layer before the last fully connected layer.
layers = [
imageInputLayer(imageInputSize,Normalization="none")
convolution2dLayer(filterSize,numFilters,Name="conv")
reluLayer
fullyConnectedLayer(50)
flattenLayer
concatenationLayer(1,2,Name="concat")
fullyConnectedLayer(numClasses)
softmaxLayer];
net = addLayers(net,layers);For the second part of the network, add a feature input layer and connect it to the second input of the concatenation layer.
featInput = featureInputLayer(numFeatures,Name="features"); net = addLayers(net,featInput); net = connectLayers(net,"features","concat/in2")
net =
dlnetwork with properties:
Layers: [9×1 nnet.cnn.layer.Layer]
Connections: [8×2 table]
Learnables: [6×3 table]
State: [0×3 table]
InputNames: {'imageinput' 'features'}
OutputNames: {'softmax'}
Initialized: 0
View summary with summary.
Visualize the network.
plot(net)
If you have a data set of numeric features (for example tabular data without spatial or time dimensions), then you can train a deep neural network using a feature input layer.
Read the transmission casing data from the CSV file "transmissionCasingData.csv".
filename = "transmissionCasingData.csv"; tbl = readtable(filename,TextType="String");
Convert the labels for prediction to categorical using the convertvars function.
labelName = "GearToothCondition"; tbl = convertvars(tbl,labelName,"categorical");
To train a network using categorical features, you must first convert the categorical features to numeric. First, convert the categorical predictors to categorical using the convertvars function by specifying a string array containing the names of all the categorical input variables. In this data set, there are two categorical features with names "SensorCondition" and "ShaftCondition".
categoricalPredictorNames = ["SensorCondition" "ShaftCondition"]; tbl = convertvars(tbl,categoricalPredictorNames,"categorical");
Loop over the categorical input variables. For each variable, convert the categorical values to one-hot encoded vectors using the onehotencode function.
for i = 1:numel(categoricalPredictorNames) name = categoricalPredictorNames(i); tbl.(name) = onehotencode(tbl.(name),2); end
View the first few rows of the table. Notice that the categorical predictors have been split into multiple columns.
head(tbl)
SigMean SigMedian SigRMS SigVar SigPeak SigPeak2Peak SigSkewness SigKurtosis SigCrestFactor SigMAD SigRangeCumSum SigCorrDimension SigApproxEntropy SigLyapExponent PeakFreq HighFreqPower EnvPower PeakSpecKurtosis SensorCondition ShaftCondition GearToothCondition
________ _________ ______ _______ _______ ____________ ___________ ___________ ______________ _______ ______________ ________________ ________________ _______________ ________ _____________ ________ ________________ _______________ ______________ __________________
-0.94876 -0.9722 1.3726 0.98387 0.81571 3.6314 -0.041525 2.2666 2.0514 0.8081 28562 1.1429 0.031581 79.931 0 6.75e-06 3.23e-07 162.13 0 1 1 0 No Tooth Fault
-0.97537 -0.98958 1.3937 0.99105 0.81571 3.6314 -0.023777 2.2598 2.0203 0.81017 29418 1.1362 0.037835 70.325 0 5.08e-08 9.16e-08 226.12 0 1 1 0 No Tooth Fault
1.0502 1.0267 1.4449 0.98491 2.8157 3.6314 -0.04162 2.2658 1.9487 0.80853 31710 1.1479 0.031565 125.19 0 6.74e-06 2.85e-07 162.13 0 1 0 1 No Tooth Fault
1.0227 1.0045 1.4288 0.99553 2.8157 3.6314 -0.016356 2.2483 1.9707 0.81324 30984 1.1472 0.032088 112.5 0 4.99e-06 2.4e-07 162.13 0 1 0 1 No Tooth Fault
1.0123 1.0024 1.4202 0.99233 2.8157 3.6314 -0.014701 2.2542 1.9826 0.81156 30661 1.1469 0.03287 108.86 0 3.62e-06 2.28e-07 230.39 0 1 0 1 No Tooth Fault
1.0275 1.0102 1.4338 1.0001 2.8157 3.6314 -0.02659 2.2439 1.9638 0.81589 31102 1.0985 0.033427 64.576 0 2.55e-06 1.65e-07 230.39 0 1 0 1 No Tooth Fault
1.0464 1.0275 1.4477 1.0011 2.8157 3.6314 -0.042849 2.2455 1.9449 0.81595 31665 1.1417 0.034159 98.838 0 1.73e-06 1.55e-07 230.39 0 1 0 1 No Tooth Fault
1.0459 1.0257 1.4402 0.98047 2.8157 3.6314 -0.035405 2.2757 1.955 0.80583 31554 1.1345 0.0353 44.223 0 1.11e-06 1.39e-07 230.39 0 1 0 1 No Tooth Fault
View the class names of the data set.
classNames = categories(tbl{:,labelName})classNames = 2×1 cell
{'No Tooth Fault'}
{'Tooth Fault' }
Set aside data for testing. Partition the data into a training set containing 85% of the data and a test set containing the remaining 15% of the data. To partition the data, use the trainingPartitions function, attached to this example as a supporting file. To access this file, open the example as a live script.
numObservations = size(tbl,1); [idxTrain,idxTest] = trainingPartitions(numObservations,[0.85 0.15]); tblTrain = tbl(idxTrain,:); tblTest = tbl(idxTest,:);
Convert the data to a format that the trainnet function supports. Convert the predictors and targets to numeric and categorical arrays, respectively. For feature input, the network expects data with rows that correspond to observations and columns that correspond to the features. If your data has a different layout, then you can preprocess your data to have this layout or you can provide layout information using data formats. For more information, see Deep Learning Data Formats.
predictorNames = ["SigMean" "SigMedian" "SigRMS" "SigVar" "SigPeak" "SigPeak2Peak" ... "SigSkewness" "SigKurtosis" "SigCrestFactor" "SigMAD" "SigRangeCumSum" ... "SigCorrDimension" "SigApproxEntropy" "SigLyapExponent" "PeakFreq" ... "HighFreqPower" "EnvPower" "PeakSpecKurtosis" "SensorCondition" "ShaftCondition"]; XTrain = table2array(tblTrain(:,predictorNames)); TTrain = tblTrain.(labelName); XTest = table2array(tblTest(:,predictorNames)); TTest = tblTest.(labelName);
Define a network with a feature input layer and specify the number of features. Also, configure the input layer to normalize the data using Z-score normalization.
numFeatures = size(XTrain,2);
numClasses = numel(classNames);
layers = [
featureInputLayer(numFeatures,Normalization="zscore")
fullyConnectedLayer(16)
layerNormalizationLayer
reluLayer
fullyConnectedLayer(numClasses)
softmaxLayer];Specify the training options:
Train using the L-BFGS solver. This solver suits tasks with small networks and when the data fits in memory.
Train using the CPU. Because the network and data is small, the CPU is better suited.
Display the training progress in a plot.
Suppress the verbose output.
options = trainingOptions("lbfgs", ... ExecutionEnvironment="cpu", ... Plots="training-progress", ... Verbose=false);
Train the network using the trainnet function. For classification, use cross-entropy loss.
net = trainnet(XTrain,TTrain,layers,"crossentropy",options);
Test the network using the labeled test set. For single-label classification, evaluate the accuracy. The accuracy is the percentage of the labels that the network predicts correctly.
accuracy = testnet(net,XTest,TTest,"accuracy")accuracy = 100
Predict the labels of the test data using the trained network. Predict the classification scores using the trained network then convert the predictions to labels using the scores2label function.
scoresTest = minibatchpredict(net,XTest); YTest = scores2label(scoresTest,classNames);
Visualize the predictions in a confusion chart.
confusionchart(TTest,YTest)

Algorithms
Layers in a layer array or layer graph pass data to subsequent layers as formatted dlarray objects.
The format of a dlarray object is a string of characters in which each
character describes the corresponding dimension of the data. The format consists of one or
more of these characters:
"S"— Spatial"C"— Channel"B"— Batch"T"— Time"U"— Unspecified
For example, you can represent tabular data as a 2-D array, in which the first and
second dimensions correspond to the batch and channel dimensions, respectively. This
representation is in the format "BC" (batch, channel).
The input layer of a network specifies the layout of the data that the network expects. If you have data in a different layout, then specify the layout using the InputDataFormats training option.
The layer inputs N-by-c arrays to the network,
where N and c are the numbers of observations and
channels of the data, respectively. Data in this layout has the data format
"BC" (batch, channel).
Extended Capabilities
Code generation does not support passing
dlarrayobjects with unspecified (U) dimensions to this layer.Code generation does not support complex input and does not support
'SplitComplexInputs'option.
To generate CUDA® or C++ code by using GPU Coder™, you must first construct and train a deep neural network. Once the network is trained and evaluated, you can configure the code generator to generate code and deploy the convolutional neural network on platforms that use NVIDIA® or ARM® GPU processors. For more information, see Deep Learning with GPU Coder (GPU Coder).
Code generation does not support passing
dlarrayobjects with unspecified (U) dimensions to this layer.Code generation does not support complex input and does not support
'SplitComplexInputs'option.
Version History
Introduced in R2020bFor complex-valued input to the neural network, when the SplitComplexIputs is 0 (false), the layer passes complex-valued data to subsequent layers.
If the input data is complex-valued and the SplitComplexInputs option is
0 (false), then the
Normalization option must be "zerocenter",
"zscore", "none", or a function handle. The
Mean and StandardDeviation properties of the layer
also support complex-valued data for the "zerocenter" and
"zscore" normalization options.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)