rrcforest
Syntax
Description
Use the rrcforest function to fit a robust random cut forest model for outlier detection and novelty
detection.
Outlier detection (detecting anomalies in training data) — Use the output argument
tfofrrcforestto identify anomalies in training data.Novelty detection (detecting anomalies in new data with uncontaminated training data) — Create a
RobustRandomCutForestmodel object by passing uncontaminated training data (data with no outliers) torrcforest. Detect anomalies in new data by passing the object and the new data to the object functionisanomaly.
forest = rrcforest(Tbl)RobustRandomCutForest model object for the predictor data in the table
Tbl.
forest = rrcforest(___,Name=Value)ContaminationFraction=0.1
Examples
Detect outliers (anomalies in training data) by using the rrcforest function.
Load the sample data set NYCHousing2015.
load NYCHousing2015The data set includes 10 variables with information on the sales of properties in New York City in 2015. Display a summary of the data set.
summary(NYCHousing2015)
NYCHousing2015: 91446×10 table
Variables:
BOROUGH: double
NEIGHBORHOOD: cell array of character vectors
BUILDINGCLASSCATEGORY: cell array of character vectors
RESIDENTIALUNITS: double
COMMERCIALUNITS: double
LANDSQUAREFEET: double
GROSSSQUAREFEET: double
YEARBUILT: double
SALEPRICE: double
SALEDATE: datetime
Statistics for applicable variables:
NumMissing Min Median Max Mean Std
BOROUGH 0 1 3 5 2.8431 1.3343
NEIGHBORHOOD 0
BUILDINGCLASSCATEGORY 0
RESIDENTIALUNITS 0 0 1 8759 2.1789 32.2738
COMMERCIALUNITS 0 0 0 612 0.2201 3.2991
LANDSQUAREFEET 0 0 1700 29305534 2.8752e+03 1.0118e+05
GROSSSQUAREFEET 0 0 1056 8942176 4.6598e+03 4.3098e+04
YEARBUILT 0 0 1939 2016 1.7951e+03 526.9998
SALEPRICE 0 0 333333 4.1111e+09 1.2364e+06 2.0130e+07
SALEDATE 0 01-Jan-2015 09-Jul-2015 31-Dec-2015 07-Jul-2015 2470:47:17
The SALEDATE column is a datetime array, which is not supported by rrcforest. Create columns for the month and day numbers of the datetime values, and then delete the SALEDATE column.
[~,NYCHousing2015.MM,NYCHousing2015.DD] = ymd(NYCHousing2015.SALEDATE); NYCHousing2015.SALEDATE = [];
The columns BOROUGH, NEIGHBORHOOD, and BUILDINGCLASSCATEGORY contain categorical predictors. Display the number of categories for the categorical predictors.
length(unique(NYCHousing2015.BOROUGH))
ans = 5
length(unique(NYCHousing2015.NEIGHBORHOOD))
ans = 254
length(unique(NYCHousing2015.BUILDINGCLASSCATEGORY))
ans = 48
For a categorical variable with more than 64 categories, the rrcforest function uses an approximate splitting method that can reduce the accuracy of the robust random cut forest model. Remove the NEIGHBORHOOD column, which contains a categorical variable with 254 categories.
NYCHousing2015.NEIGHBORHOOD = [];
Train a robust random cut forest model for NYCHousing2015. Specify the fraction of anomalies in the training observations as 0.1, and specify the first variable (BOROUGH) as a categorical predictor. The first variable is a numeric array, so rrcforest assumes it is a continuous variable unless you specify the variable as a categorical variable.
rng("default") % For reproducibility [Mdl,tf,scores] = rrcforest(NYCHousing2015, ... ContaminationFraction=0.1,CategoricalPredictors=1);
Mdl is a RobustRandomCutForest model object. rrcforest also returns the anomaly indicators (tf) and anomaly scores (scores) for the training data NYCHousing2015.
Plot a histogram of the score values. Create a vertical line at the score threshold corresponding to the specified fraction.
histogram(scores) xline(Mdl.ScoreThreshold,"r-",["Threshold" Mdl.ScoreThreshold])

If you want to identify anomalies with a different contamination fraction (for example, 0.01), you can train a new robust random cut forest model.
rng("default") % For reproducibility [newMdl,newtf,scores] = rrcforest(NYCHousing2015, ... ContaminationFraction=0.01,CategoricalPredictors=1);
If you want to identify anomalies with a different score threshold value (for example, 65), you can pass the RobustRandomCutForest model object, the training data, and a new threshold value to the isanomaly function.
[newtf,scores] = isanomaly(Mdl,NYCHousing2015,ScoreThreshold=65);
Note that changing the contamination fraction or score threshold changes the anomaly indicators only, and does not affect the anomaly scores. Therefore, if you do not want to compute the anomaly scores again by using rrcforest or isanomaly, you can obtain a new anomaly indicator using the existing score values.
Change the fraction of anomalies in the training data to 0.01.
newContaminationFraction = 0.01;
Find a new score threshold by using the quantile function.
newScoreThreshold = quantile(scores,1-newContaminationFraction)
newScoreThreshold = 63.2642
Obtain a new anomaly indicator.
newtf = scores > newScoreThreshold;
Create a RobustRandomCutForest model object for uncontaminated training observations by using the rrcforest function. Then detect novelties (anomalies in new data) by passing the object and the new data to the object function isanomaly.
Load the 1994 census data stored in census1994.mat. The data set contains demographic data from the US Census Bureau to predict whether an individual makes over $50,000 per year.
load census1994census1994 contains the training data set adultdata and the test data set adulttest.
Assume that adultdata does not contain outliers. Train a robust random cut forest model for adultdata. Specify StandardizeData as true to standardize the input data.
rng("default") % For reproducibility [Mdl,tf,s] = rrcforest(adultdata,StandardizeData=true);
Mdl is a RobustRandomCutForest model object. rrcforest also returns the anomaly indicators tf and anomaly scores s for the training data adultdata. If you do not specify the ContaminationFraction name-value argument as a value greater than 0, then rrcforest treats all training observations as normal observations, meaning all the values in tf are logical 0 (false). The function sets the score threshold to the maximum score value. Display the threshold value.
Mdl.ScoreThreshold
ans = 86.5315
Find anomalies in adulttest by using the trained robust random cut forest model. Because you specified StandardizeData=true when you trained the model, the isanomaly function standardizes the input data by using the predictor means and standard deviations of the training data stored in the Mu and Sigma properties, respectively.
[tf_test,s_test] = isanomaly(Mdl,adulttest);
The isanomaly function returns the anomaly indicators tf_test and scores s_test for adulttest. By default, isanomaly identifies observations with scores above the threshold (Mdl.ScoreThreshold) as anomalies.
Create histograms for the anomaly scores s and s_test. Create a vertical line at the threshold of the anomaly scores.
histogram(s,Normalization="probability") hold on histogram(s_test,Normalization="probability") xline(Mdl.ScoreThreshold,"r-",join(["Threshold" Mdl.ScoreThreshold])) legend("Training Data","Test Data",Location="northwest") hold off
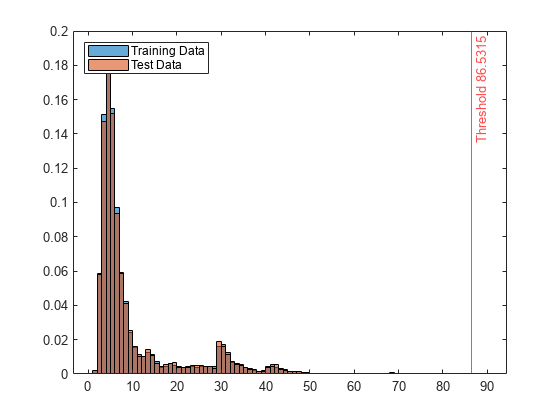
Display the observation index of the anomalies in the test data.
find(tf_test)
ans = 3541
The anomaly score distribution of the test data is similar to that of the training data, so isanomaly detects a small number of anomalies in the test data with the default threshold value.
Zoom in to see the anomaly and the observations near the threshold.
xlim([50 92]) ylim([0 0.001])
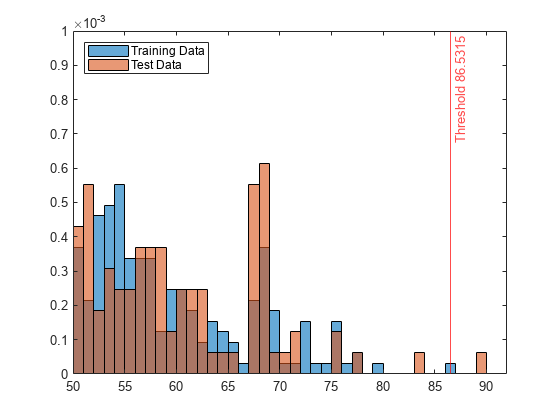
You can specify a different threshold value by using the ScoreThreshold name-value argument. For an example, see Specify Anomaly Score Threshold.
Input Arguments
Predictor data, specified as a table. Each row of Tbl corresponds to one observation, and each column corresponds to one predictor variable. Multicolumn variables and cell arrays other than cell arrays of character vectors are not allowed.
To use a subset of the variables in Tbl, specify the variables by using the PredictorNames name-value argument.
Data Types: table
Predictor data, specified as a numeric matrix. Each row of X corresponds to one observation, and each column corresponds to one predictor variable.
You can use the PredictorNames name-value argument to assign names to the predictor variables in X.
Data Types: single | double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: NumLearners=50,NumObservationsPerLearner=100 specifies to
train a robust random cut forest model using 50 trees and 100 observations for each
tree.
List of categorical predictors, specified as one of the values in this table.
| Value | Description |
|---|---|
| Vector of positive integers | Each entry in the vector is an index value indicating that the corresponding predictor is categorical. The index values are between 1 and If |
| Logical vector | A |
| Character matrix | Each row of the matrix is the name of a predictor variable. The names must match the entries
in PredictorNames. Pad the names
with extra blanks so each row of the character matrix has the same
length. |
| String array or cell array of character vectors | Each element in the array is the name of a predictor variable. The names must match the entries in PredictorNames. |
"all" | All predictors are categorical. |
By default, if the predictor data is a table
(Tbl), rrcforest assumes that a variable is
categorical if it is a logical vector, unordered categorical vector, character array, string
array, or cell array of character vectors. If the predictor data is a matrix
(X), rrcforest assumes that all predictors are
continuous. To identify any other predictors as categorical predictors, specify them by using
the CategoricalPredictors name-value argument.
For a categorical variable with more than 64 categories, the
rrcforest function uses an approximate splitting method that can
reduce the accuracy of the model.
Example: CategoricalPredictors="all"
Data Types: single | double | logical | char | string | cell
Collusive displacement calculation method, specified as
"maximal" or "average".
The rrcforest function finds the maximum change
("maximal") or the average change ("average")
in model complexity for each tree, and computes the collusive displacement (anomaly
score) for each observation. For details, see Anomaly Scores.
Example: CollusiveDisplacement="average"
Data Types: char | string
Fraction of anomalies in the training data, specified as a numeric scalar in the range [0,1].
If the
ContaminationFractionvalue is 0 (default), thenrrcforesttreats all training observations as normal observations, and sets the score threshold (ScoreThresholdproperty value offorest) to the maximum value ofscores.If the
ContaminationFractionvalue is in the range (0,1], thenrrcforestdetermines the threshold value so that the function detects the specified fraction of training observations as anomalies.
Example: ContaminationFraction=0.1
Data Types: single | double
Number of robust random cut trees (trees in the robust random cut forest model), specified as a positive integer scalar.
Example: NumLearners=50
Data Types: single | double
Number of observations to draw from the training data without replacement for each robust random cut tree (tree in the robust random cut forest model), specified as a positive integer scalar greater than or equal to 3.
Example: NumObservationsPerLearner=100
Data Types: single | double
This property is read-only.
Predictor variable names, specified as a string array of unique names or cell array of
unique character vectors. The functionality of PredictorNames depends
on how you supply the predictor data.
If you supply
Tbl, then you can usePredictorNamesto specify which predictor variables to use. That is,rrcforestuses only the predictor variables inPredictorNames.PredictorNamesmust be a subset ofTbl.Properties.VariableNames.By default,
PredictorNamescontains the names of all predictor variables inTbl.
If you supply
X, then you can usePredictorNamesto assign names to the predictor variables inX.The order of the names in
PredictorNamesmust correspond to the column order ofX. That is,PredictorNames{1}is the name ofX(:,1),PredictorNames{2}is the name ofX(:,2), and so on. Also,size(X,2)andnumel(PredictorNames)must be equal.By default,
PredictorNamesis{"x1","x2",...}.
Data Types: string | cell
Flag to standardize the predictor data, specified as a numeric or logical 1
(true) or 0 (false).
If you set StandardizeData=true, the rrcforest function centers and scales each predictor variable (X or Tbl) by the corresponding column mean and standard deviation. The function does not standardize the data contained in the dummy variable columns generated for categorical predictors.
Example: StandardizeData=true
Data Types: logical
Flag to run in parallel, specified as a numeric or logical 1
(true) or 0 (false). If you specify
UseParallel=true, the rrcforest function executes
for-loop iterations by using parfor. The loop runs in parallel when you have Parallel Computing Toolbox™.
Example: UseParallel=true
Data Types: logical
Output Arguments
Trained robust random cut forest model, returned as a RobustRandomCutForest model object.
You can use the object function isanomaly
with forest to find anomalies in new data.
Anomaly indicators, returned as a logical column vector. An element of
tf is true when the observation in the
corresponding row of Tbl or X is an anomaly,
and false otherwise. tf has the same length as
Tbl or X.
rrcforest identifies observations with
scores above the threshold (ScoreThreshold property value of forest) as
anomalies. The function determines the threshold value to detect the specified fraction
(ContaminationFraction name-value argument) of training
observations as anomalies.
Anomaly scores, returned as a numeric column vector with values in the range [0,Inf). scores has the same length as
Tbl or X, and each element of
scores contains an anomaly score for the observation in the
corresponding row of Tbl or X. A small
positive value indicates a normal observation, and a large positive value indicates an
anomaly.
More About
The robust random cut forest algorithm [1] classifies a point as a normal point or an anomaly based on the change in model complexity introduced by the point. Similar to the Isolation Forest algorithm, the robust random cut forest algorithm builds an ensemble of trees. The two algorithms differ in how they choose a split variable in the trees and how they define anomaly scores.
The rrcforest function creates a robust random cut forest model (ensemble
of robust random cut trees) for training observations and detects outliers (anomalies in the
training data). Each tree is trained for a subset of training observations as follows:
rrcforestdraws samples without replacement from the training observations for each tree.rrcforestgrows a tree by choosing a split variable in proportion to the ranges of variables, and choosing the split position uniformly at random. The function continues until every sample reaches a separate leaf node for each tree.
Using the range information in to choose a split variable makes the algorithm robust to irrelevant variables.
Anomalies are easy to describe, but make describing the remainder of the data more
difficult. Therefore, adding an anomaly to a model increases the model complexity of a
forest model [1]. The rrcforest
function identifies outliers using anomaly scores that are defined
based on the change in model complexity.
The isanomaly function uses a trained robust random cut forest model to
detect anomalies in the data. For novelty detection (detecting anomalies in new data with
uncontaminated training data), you can train a robust random cut forest model with
uncontaminated training data (data with no outliers) and use it to detect anomalies in new
data. For each observation of the new data, the function finds the corresponding leaf node
in each tree, computes the change in model complexity introduced by the leaf nodes, and
returns an anomaly indicator and score.
The robust random cut forest algorithm uses collusive displacement as an anomaly score. The collusive displacement of a point x indicates the contribution of x to the model complexity of a forest model. A small positive anomaly score value indicates a normal observation, and a large positive value indicates an anomaly.
As defined in [1], the model complexity |M(T)| of a tree T is the sum of path lengths (the distance from the root node to the leaf nodes) over all points in the training data Z.
where f(y,Z,T) is the depth of y in tree T. The displacement of x is defined to indicate the expected changes in the model complexity introduced by x.
where T' is a tree over Z – {x}. Disp(x,Z) is the expected number of points in the sibling node of the leaf node
containing x. This definition is not robust to duplicates or
near-duplicates, and can cause outlier masking. To avoid outlier masking, the robust random
cut forest algorithm uses the collusive displacement CoDisp, where a set
C includes x and the colluders of
x.
where T" is a tree over Z – C, and |C| is the number of points in the subtree of T for C.
The default value for the CollusiveDisplacement name-value argument of rrcforest
is "maximal". For each tree, by default, the software finds the set
C that maximizes the ratio Disp(x,C)/|C| by traversing from the leaf node of x to the root node,
as described in [2]. If you specify
CollusiveDisplacement="average"
Algorithms
rrcforest considers NaN, '' (empty character vector), "" (empty string), <missing>, and <undefined> values in Tbl and NaN values in X to be missing values.
rrcforest uses observations with missing values to find splits on
variables for which these observations have valid values. The function might place these
observations in a branch node, not a leaf node. Then rrcforest
computes the ratio (Disp(x,C)/|C|) by traversing from the branch node to the root node for each tree. The
function places an observation with all missing values in the root node. Therefore, the
ratio and the anomaly score become the number of training observations for each tree, which
is the maximum possible anomaly score for the trained robust random cut forest model. You
can specify the number of training observations for each tree by using the NumObservationsPerLearner name-value argument.
References
[1] Guha, Sudipto, N. Mishra, G. Roy, and O. Schrijvers. "Robust Random Cut Forest Based Anomaly Detection on Streams," Proceedings of The 33rd International Conference on Machine Learning 48 (June 2016): 2712–21.
[2] Bartos, Matthew D., A. Mullapudi, and S. C. Troutman. "rrcf: Implementation of the Robust Random Cut Forest Algorithm for Anomaly Detection on Streams." Journal of Open Source Software 4, no. 35 (2019): 1336.
Extended Capabilities
To run in parallel, set the UseParallel name-value argument to
true in the call to this function.
For more general information about parallel computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
Version History
Introduced in R2023a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)