knntest
Two-sample multivariate hypothesis test using k-nearest neighbors (KNN)
Since R2025a
Syntax
Description
knnstat = knntest(X,Y)knnstat for the multivariate data
sets X and Y. The statistic indicates how well
separated the X and Y data sets are, based on
whether the observations' nearest neighbors tend to be in the same set as the observations.
For more information, see Nearest Neighbor Statistic.
knnstat = knntest(X,Y,Name=Value)
[
also returns the p-value knnstat,p] = knntest(___)p of the hypothesis test,
using any of the input argument combinations in the previous syntaxes. For more information,
see p-Value Computation and Hypothesis Test.
Examples
Calculate and compare the nearest neighbor statistic values for cars manufactured in three different years to determine which two years have the most similar distribution of automobile measurements.
Load the carsmall data set, which contains measurements of cars manufactured in 1970, 1976, and 1982. Create a table from this data and display the first eight rows.
load carsmall carData = table(Acceleration,Cylinders,Displacement, ... Horsepower,Model_Year,Origin,MPG,Weight); head(carData)
Acceleration Cylinders Displacement Horsepower Model_Year Origin MPG Weight
____________ _________ ____________ __________ __________ _______ ___ ______
12 8 307 130 70 USA 18 3504
11.5 8 350 165 70 USA 15 3693
11 8 318 150 70 USA 18 3436
12 8 304 150 70 USA 16 3433
10.5 8 302 140 70 USA 17 3449
10 8 429 198 70 USA 15 4341
9 8 454 220 70 USA 14 4354
8.5 8 440 215 70 USA 14 4312
Display the number of cars manufactured in each year.
groupsummary(carData,"Model_Year")ans=3×2 table
Model_Year GroupCount
__________ __________
70 35
76 34
82 31
The data set contains a similar number of cars for each year in Model_Year.
Create separate tables containing all the data for cars manufactured in 1970, 1976, and 1982.
car70 = carData(carData.Model_Year==70,:); car76 = carData(carData.Model_Year==76,:); car82 = carData(carData.Model_Year==82,:);
Create a vector containing the names of all the variables except Model_Year. Because the data sets have different values for Model_Year, omit this variable from the nearest neighbor statistic computation.
variableNames = ["Acceleration","Cylinders","Displacement", ... "Horsepower","Origin","MPG","Weight"];
Use the knntest function to calculate the nearest neighbor statistic for the 1970 and 1976 data sets, the 1976 and 1982 data sets, and the 1970 and 1982 data sets. Specify which variables to include in the computation by using the VariableNames name-value argument.
knn7076 = knntest(car70,car76,VariableNames=variableNames); knn7682 = knntest(car76,car82,VariableNames=variableNames); knn7082 = knntest(car70,car82,VariableNames=variableNames);
Display the three nearest neighbor statistic values in a bar graph. Recall that the nearest neighbor statistic indicates how well separated two data sets are, based on whether the observations' nearest neighbors tend to be in the same set as the observations. In general, a value closer to 1 indicates greater dissimilarity between two data sets.
years = ["1970 & 1976","1976 & 1982","1970 & 1982"]; knnValues = [knn7076,knn7682,knn7082]; bar(years,knnValues) ylabel("KNN Values")

The bar graph shows that 1970 and 1976 have the smallest nearest neighbor statistic value. This result indicates that 1970 and 1976 have the most similar distribution of car measurements.
Perform a two-sample hypothesis test using a nearest neighbor statistic to determine if two iris species have the same distribution of sepal and petal dimensions. The null hypothesis of the test is that the data sets for the two iris species come from the same distribution. The alternative hypothesis is that the data sets come from different distributions.
First, perform a hypothesis test on two samples of iris data with equal numbers of each iris species. Load the fisheriris data set into a table and display the first eight rows.
fisheriris = readtable("fisheriris.csv");
head(fisheriris) SepalLength SepalWidth PetalLength PetalWidth Species
___________ __________ ___________ __________ __________
5.1 3.5 1.4 0.2 {'setosa'}
4.9 3 1.4 0.2 {'setosa'}
4.7 3.2 1.3 0.2 {'setosa'}
4.6 3.1 1.5 0.2 {'setosa'}
5 3.6 1.4 0.2 {'setosa'}
5.4 3.9 1.7 0.4 {'setosa'}
4.6 3.4 1.4 0.3 {'setosa'}
5 3.4 1.5 0.2 {'setosa'}
Split the data set into two samples with even distribution of the species.
cv = cvpartition(fisheriris.Species,"Holdout",0.5);
sample1 = fisheriris(cv.training,:);
sample2 = fisheriris(cv.test,:);Perform a hypothesis test at the 1% significance level using the knntest function.
[knnValue,p,h] = knntest(sample1,sample2,Alpha=0.01)
knnValue = 0.5167
p = 0.1364
h = 0
The returned test decision of h = 0 indicates that knntest fails to reject the null hypothesis that the samples come from the same distribution at the 1% significance level. The value of knnValue is close to 0.5, which suggests that the samples have similar distributions.
Next, perform a hypothesis test to compare the distribution of petal and sepal data for the setosa and virginica iris species. Create separate tables containing the data for the setosa and virginica iris species.
setosa = fisheriris(string(fisheriris.Species)=="setosa",:); virginica = fisheriris(string(fisheriris.Species)=="virginica",:);
Store the sepal and petal data for each species in a numeric matrix.
setosaData = setosa{:,1:end-1};
virginicaData = virginica{:,1:end-1};Perform a hypothesis test at the 1% significance level using the knntest function.
[knnValue2,p2,h2] = knntest(setosaData,virginicaData,Alpha=0.01)
knnValue2 = 1
p2 = 2.9384e-113
h2 = 1
The returned test decision of h = 1 indicates that knntest rejects the null hypothesis that the samples come from the same distribution at the 1% significance level. This result indicates that the setosa and virginica iris species have different distributions of sepal and petal data.
Evaluate data synthesized from an existing data set. Compare the existing and synthetic data sets to determine distribution similarity.
Load the carsmall data set, which contains measurements of cars manufactured in 1970, 1976, and 1982. Create a table containing the data and display the first eight observations.
load carsmall carData = table(Acceleration,Cylinders,Displacement,Horsepower, ... Mfg,Model,Model_Year,MPG,Origin,Weight); head(carData)
Acceleration Cylinders Displacement Horsepower Mfg Model Model_Year MPG Origin Weight
____________ _________ ____________ __________ _____________ _________________________________ __________ ___ _______ ______
12 8 307 130 chevrolet chevrolet chevelle malibu 70 18 USA 3504
11.5 8 350 165 buick buick skylark 320 70 15 USA 3693
11 8 318 150 plymouth plymouth satellite 70 18 USA 3436
12 8 304 150 amc amc rebel sst 70 16 USA 3433
10.5 8 302 140 ford ford torino 70 17 USA 3449
10 8 429 198 ford ford galaxie 500 70 15 USA 4341
9 8 454 220 chevrolet chevrolet impala 70 14 USA 4354
8.5 8 440 215 plymouth plymouth fury iii 70 14 USA 4312
Generate 100 new observations using the synthesizeTabularData function. Specify the Cylinders and Model_Year variables as discrete numeric variables. Display the first eight observations.
rng(0,"twister") syntheticData = synthesizeTabularData(carData,100, ... DiscreteNumericVariables=["Cylinders","Model_Year"]); head(syntheticData)
Acceleration Cylinders Displacement Horsepower Mfg Model Model_Year MPG Origin Weight
____________ _________ ____________ __________ _____________ _________________________________ __________ ______ _______ ______
11.215 8 309.73 137.28 dodge dodge coronet brougham 76 17.3 USA 4038
10.198 8 416.68 215.51 plymouth plymouth fury iii 70 9.5497 USA 4507.2
17.161 6 258.38 77.099 amc amc pacer d/l 76 18.325 USA 3199.8
9.4623 8 426.19 197.3 plymouth plymouth fury iii 70 11.747 USA 4372.1
13.992 4 106.63 91.396 datsun datsun pl510 70 30.56 Japan 1950.7
17.965 6 266.24 78.719 oldsmobile oldsmobile cutlass ciera (diesel) 82 36.416 USA 2832.4
17.028 4 139.02 100.24 chevrolet chevrolet cavalier 2-door 82 36.058 USA 2744.5
15.343 4 118.93 100.22 toyota toyota celica gt 82 26.696 Japan 2600.5
Visualize the synthetic and existing data sets. Create a DriftDiagnostics object using the detectdrift function. The object's plotEmpiricalCDF and plotHistogram functions let you visualize continuous and discrete variables.
dd = detectdrift(carData,syntheticData);
Use plotEmpiricalCDF to visualize the empirical cumulative distribution function (ECDF) of the values in carData and syntheticData.
continuousVariable ="Acceleration"; plotEmpiricalCDF(dd,Variable=continuousVariable) legend(["Existing data","Synthetic data"])
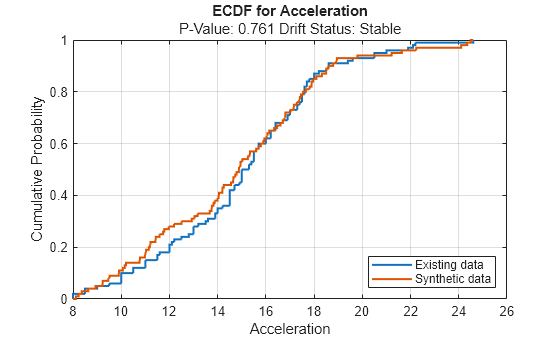
For the variable Acceleration, the ECDF of the existing data (in blue) and the ECDF of the synthetic data (in red) appear to be similar.
Use plotHistogram to visualize the distribution of values for the discrete variables in carData and syntheticData.
discreteVariable ="Cylinders"; plotHistogram(dd,Variable=discreteVariable) legend(["Existing data","Synthetic data"])
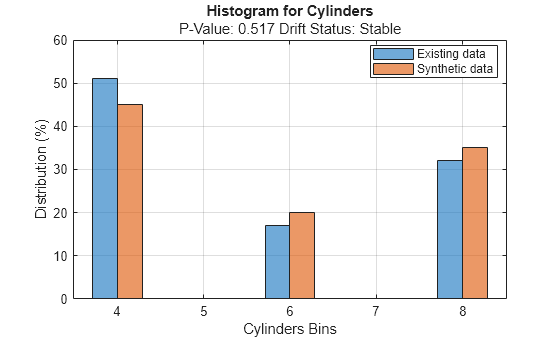
For the variable Cylinders, the distribution of data between the bins for the existing data (in blue) and the synthetic data (in red) appear similar.
Compare the synthetic and existing data sets using the knntest function. The function performs a two-sample hypothesis test for the null hypothesis that the samples come from the same distribution.
[knnstat,p,h] = knntest(carData,syntheticData)
knnstat = 0.4933
p = 0.6172
h = 0
The returned value of h = 0 indicates that knntest fails to reject the null hypothesis that the samples come from different distributions at the 5% significance level. As with other hypothesis tests, this result does not guarantee that the null hypothesis is true. That is, the samples do not necessarily come from the same distribution, but both the knnstat value close to 0.5 and the high p-value indicate that the distributions of the existing and synthetic data sets are similar.
Input Arguments
Sample data, specified as a numeric matrix or a table. The rows of
X correspond to observations, and the columns correspond to
variables. knntest ignores observations with missing data.
If
XandYare numeric matrices, they must have the same number of variables, but can have different numbers of observations.If
XandYare tables, they must have the same variable names, or the variable names of one must be a subset of the other.XandYcan have different numbers of observations.
knntest uses X as reference data to
normalize continuous variables and establish a set of categories for categorical
variables. When a categorical variable is in both X and
Y, the variable in Y cannot contain
categories that are not in X.
Data Types: single | double | table
Sample data, specified as a numeric matrix or a table. The rows of
Y correspond to observations, and the columns correspond to
variables. knntest ignores observations with missing data.
If
XandYare numeric matrices, they must have the same number of variables, but can have different numbers of observations.If
XandYare tables, they must have the same variable names, or the variable names of one must be a subset of the other.XandYcan have different numbers of observations.
Data Types: single | double | table
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: knntest(X,Y,NumNeighbors=15,Distance="euclidean") specifies a
test that finds the 15 nearest neighbors for each observation in X and
Y by using the Euclidean distance.
Significance level of the hypothesis test, specified as a scalar value in the range (0,1).
Example: Alpha=0.01
Data Types: single | double
Number of nearest neighbors to use in the computation of the statistic
knnstat, specified as a positive integer scalar.
knntest uses either a Kd-tree algorithm or an exhaustive search algorithm to find the nearest
neighbors. If knntest uses 10 or fewer variables in
X and Y, all of which are continuous, and
the Distance value is "cityblock" or
"euclidean", then the function finds nearest neighbors by using a
Kd-tree. Otherwise, the function uses an
exhaustive search algorithm.
Example: NumNeighbors=25
Data Types: single | double
Distance metric, specified as a character vector or string scalar.
If all the variables are continuous (numeric), then you can specify one of these distance metrics.
Value Description "cityblock"City block distance
"correlation"One minus the sample correlation between observations (treated as sequences of values)
"cosine"One minus the cosine of the included angle between observations (treated as vectors)
"euclidean"Euclidean distance
"fasteuclidean"Euclidean distance computed by using an alternative algorithm that saves time when the number of variables is at least 10. In some cases, this faster algorithm can reduce accuracy. "seuclidean"Standardized Euclidean distance. Each coordinate difference between observations is scaled by dividing by the corresponding element of the standard deviation
S = std([X Y],"omitnan")."fastseuclidean"Standardized Euclidean distance computed by using an alternative algorithm that saves time when the number of variables is at least 10. In some cases, this faster algorithm can reduce accuracy. Note
If you specify one of these distance metrics and the data includes categorical variables, then the software treats each categorical variable as a numeric variable for the distance computation, with each category represented by a positive integer.
If all the variables are categorical, then you can specify the following distance metric.
Value Description "hamming"Hamming distance, which is the percentage of coordinates that differ
Note
If you specify this distance metric and the data includes continuous (numeric) variables, then the software treats each continuous variable as a categorical variable for the distance computation.
If the variables are a mix of continuous (numeric) and categorical variables, then you can specify the following distance metric.
Value Description "goodall3"Modified Goodall distance
The default value is "seuclidean" if all the variables are
continuous, "hamming" if all the variables are categorical, and
"goodall3" if the variables are a mix of continuous and
categorical variables. For more information on the various distance metrics, see Distance Metrics.
Example: Distance="euclidean"
Data Types: char | string
Variables to include in the computation, specified as a character vector, string
array, or cell array of character vectors. Because matrices do not have named
variables, this argument applies only when X and
Y are tables.
VariableNamesmust be a subset of the variables shared byXandY.By default,
VariableNamescontains all the variables shared byXandY.
Example: VariableNames=["Name","Age","Score"]
Data Types: char | string | cell
Variables to treat as categorical, specified as one of the values in this table.
| Value | Description |
|---|---|
"all" | All variables contain categorical values. |
| Vector of numeric indices | Each vector element corresponds to the index of a variable, indicating that the variable contains categorical values. |
| Logical vector | A logical vector the same length as VariableNames. A
value of true indicates that the corresponding variable
contains categorical values. |
| String array or cell array of character vectors | An array containing the names of variables with categorical values. The
array elements must be names found in
VariableNames. |
If X and Y are numeric matrices, then
CategoricalVariables cannot be a string array or cell array of
character vectors. By default, knntest assumes all variables in
numeric matrices are continuous, and treats table columns of character arrays or
string scalars as categorical variables.
Example: CategoricalVariables="all"
Data Types: single | double | logical | char | string | cell
Output Arguments
Nearest neighbor statistic, returned as a scalar value in the range [0,1]. If
X and Y are well separated, then the
knnstat value is close to 1. If X and
Y are similar data sets with the same number of observations,
then the knnstat value tends to be close to 0.5. For more
information, see Nearest Neighbor Statistic.
p-value of the test, returned as a scalar value in the range [0,1].
p is the probability of observing a test statistic that is as
extreme as, or more extreme than, the observed value under the null hypothesis. A small
value of p indicates that the null hypothesis might not be
valid.
Hypothesis test result, returned as 1 or 0.
A value of
1indicates the rejection of the null hypothesis at theAlphasignificance level.A value of
0indicates a failure to reject the null hypothesis at theAlphasignificance level.
More About
A distance metric is a function that defines a
distance between two observations. knntest supports various
distance metrics for continuous (numeric) variables, categorical variables, and a mix of
continuous and categorical variables. To specify the distance metric to use for computing
the nearest neighbor statistic, set the Distance
name-value argument of knntest.
Given an mx-by-n data matrix X, which is treated as mx (1-by-n) row vectors x1, x2, ..., xmx, and an my-by-n data matrix Y, which is treated as my (1-by-n) row vectors y1, y2, ...,ymy, the various distances between the vector xs and yt are defined as follows:
City block distance
The city block distance is a special case of the Minkowski distance, where p = 1.
Correlation distance
where
and
Cosine distance
Euclidean distance
The Euclidean distance is a special case of the Minkowski distance, where p = 2.
Fast Euclidean distance
The fast Euclidean distance is the same as the Euclidean distance, but is computed using an alternative algorithm that saves time when the number of variables is at least 10. In some cases, this faster algorithm can reduce accuracy. For more information, see Fast Euclidean Distance Algorithm.
Hamming distance
The Hamming distance is the percentage of coordinates that differ.
Modified Goodall distance
The modified Goodall distance is a variant of the Goodall distance, which assigns a small distance if the matching values are infrequent regardless of the frequencies of the other values. For mismatches, the distance contribution of the variable is 1/(number of variables).
Standardized Euclidean distance
where V is the n-by-n diagonal matrix whose jth diagonal element is (S(j))2, where S is a vector of scaling factors for each dimension.
Fast standardized Euclidean distance
The fast standardized Euclidean distance is the same as the standardized Euclidean distance, but is computed using an alternative algorithm that saves time when the number of variables is at least 10. In some cases, this faster algorithm can reduce accuracy. For more information, see Fast Euclidean Distance Algorithm.
Tips
Avoid specifying small values for
NumNeighbors(such asNumNeighbors=1orNumNeighbors=2).knntestassumes that the number of nearest neighbors, the number of observations inXandY, and the number of variables inXandYare sufficiently large. For more information, see [1].
Algorithms
The nearest neighbor statistic knnstat indicates how well separated
the X and Y data sets are, based on whether the
observations' nearest neighbors tend to be in the same set as the observations.
Suppose m is the total number of observations in
X and Y combined. Define an
m-by-NumNeighbors matrix I,
where each row corresponds to an observation in X or
Y, and each column corresponds to a nearest neighbor. Let
I(i,j) = 1 if observation i and its nearest neighbor
j are in the same data set, and I(i,j) = 0
otherwise. The statistic knnstat is equal to
mean(I,"all").
If X and Y are well separated, then the
knnstat value is close to 1. If X and
Y are similar data sets with the same number of observations, then
the knnstat value tends to be close to 0.5. For more information, see
[1].
To determine p and h, the
knntest function uses a right-tailed hypothesis test that assumes the
test statistic (knnstat –
μ)/σ has a standard normal distribution.
μ is defined as:
mx is the number of observations in
X.my is the number of observations in
Y.m is the total number of observations in
XandY.
σ2 is defined as:
k is the number of nearest neighbors specified by the
NumNeighborsname-value argument.
The normal distribution assumption is based on the limiting behavior of the
test statistic as k, m, and the number of variables in
X and Y increase. For more information, see
[1].
References
[1] Williams, M. “How Good Are Your Fits? Unbinned Multivariate Goodness-of-Fit Tests in High Energy Physics.” Journal of Instrumentation 5, no. 09 (September 9, 2010): P09004. https://doi.org/10.1088/1748-0221/5/09/P09004.
Version History
Introduced in R2025a
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)