fairnessMetrics
Description
fairnessMetrics computes fairness metrics (bias and group
metrics) for a data set or binary classification model with respect to sensitive attributes.
The data-level evaluation examines binary, true labels of the data. The model-level evaluation
examines the predicted labels returned by one or more binary classification models, using both
true labels and predicted labels.
Bias metrics measure differences across groups, and group metrics contain information within the group. You can use the metrics to determine if your data or models contain bias toward a group within each sensitive attribute.
After creating a fairnessMetrics object, use the report function to
generate a fairness metrics report or use the plot function to
create a bar graph of the metrics.
Creation
Syntax
Description
metricsResults = fairnessMetrics(SensitiveAttributes,Y)Y with respect to the sensitive attributes in the
SensitiveAttributes matrix. The fairnessMetrics
function returns the fairnessMetrics object
metricsResults, which stores bias metrics and group metrics in the
BiasMetrics and
GroupMetrics
properties, respectively.
metricsResults = fairnessMetrics(Tbl,ResponseName)Tbl. The input argument ResponseName
specifies the name of the variable in Tbl that contains the class
labels.
metricsResults = fairnessMetrics(___,SensitiveAttributeNames=sensitiveAttributeNames)Tbl (whose names correspond to
sensitiveAttributeNames) as sensitive attributes, or assigns names
to the sensitive attributes in sensitiveAttributeNames. You can
specify this argument in addition to any of the input argument combinations in the
previous syntaxes.
metricsResults = fairnessMetrics(___,Predictions=predictions)predictions argument.
fairnessMetrics uses both true labels and predicted labels for the
model-level evaluation.
metricsResults = fairnessMetrics(___,Name=Value)SensitiveAttributeNames="age",ReferenceGroup=30 to compute bias
metrics for each group in the age variable with respect to the
reference age group 30.
Input Arguments
Sensitive attributes, specified as a vector or matrix. If you specify
SensitiveAttributes as a matrix, each row of
SensitiveAttributes corresponds to one observation, and each
column corresponds to one sensitive attribute.
You can use the sensitiveAttributeNames argument to assign names to the variables in
SensitiveAttributes.
Data Types: single | double | logical | char | string | cell | categorical
True, binary class labels, specified as a categorical, character, or string array; a logical or numeric vector; or a cell array of character vectors.
fairnessMetricssupports only binary classification.Ymust contain exactly two distinct classes.You can specify one of the two classes as a positive class by using the
PositiveClassname-value argument.The length of
Ymust be equal to the number of observations inSensitiveAttributesorTbl.If
Yis a character array, then each label must correspond to one row of the array.
Data Types: single | double | logical | char | string | cell | categorical
Sample data, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one sensitive
attribute. Multicolumn variables and cell arrays other than cell arrays of character
vectors are not allowed.
Optionally, Tbl can contain columns for the true class
labels, predicted class labels, and observation weights.
You must specify the true class label variable using
ResponseName, the predicted class label variables usingPredictions, and the observation weight variable usingWeights.fairnessMetricsuses the remaining variables as sensitive attributes. To use a subset of the remaining variables inTblas sensitive attributes, specify the variables by usingsensitiveAttributeNames.The true class label variable must be a categorical, character, or string array, a logical or numeric vector, or a cell array of character vectors.
fairnessMetricssupports only binary classification. The true class label variable must contain exactly two distinct classes.You can specify one of the two classes as a positive class by using the
PositiveClassname-value argument.
The column for the weights must be a numeric vector.
If Tbl does not contain the true class label variable, then
specify the variable by using Y. The
length of the response variable Y and the number of rows in
Tbl must be equal. To use a subset of the variables in
Tbl as sensitive attributes, specify the variables by using
sensitiveAttributeNames.
Data Types: table
Name of the true class label variable, specified as a character vector or string
scalar containing the name of the response variable in Tbl.
Example: "trueLabel" indicates that the
trueLabel variable in Tbl
(Tbl.trueLabel) is the true class label variable.
Data Types: char | string
Names of the sensitive attribute variables, specified as a character vector,
string array of unique names, or cell array of unique character vectors. The
functionality of sensitiveAttributeNames depends on the way you
supply the sample data.
If you supply
SensitiveAttributesandY, then you can usesensitiveAttributeNamesto assign names to the variables inSensitiveAttributes.The order of the names in
sensitiveAttributeNamesmust correspond to the column order ofSensitiveAttributes. That is,sensitiveAttributeNames{1}is the name ofSensitiveAttributes(:,1),sensitiveAttributeNames{2}is the name ofSensitiveAttributes(:,2), and so on. Also,size(SensitiveAttributes,2)andnumel(sensitiveAttributeNames)must be equal.By default,
sensitiveAttributeNamesis{'x1','x2',...}.
If you supply
Tbl, then you can usesensitiveAttributeNamesto specify the variables to use as sensitive attributes. That is,fairnessMetricsuses only the variables insensitiveAttributeNamesto compute fairness metrics.sensitiveAttributeNamesmust be a subset ofTbl.Properties.VariableNamesand cannot include the name of a class label variable or observation weight variable.By default,
sensitiveAttributeNamesis a set of all variable names inTbl, except the variables specified byResponseName,Predictions, andWeights.
Example: SensitiveAttributeNames="Gender"
Example: SensitiveAttributeNames=["age","marital_status"]
Data Types: char | string | cell
Predicted class labels (model predictions), specified as [],
the names of variables in Tbl, or a matrix.
Before R2023a: Specify predicted labels for at most one
binary classifier by using the name of a variable in Tbl or a
vector.
[]—fairnessMetricscomputes fairness metrics for the true class label variable (Yor theResponseNamevariable inTbl).Names of variables in
Tbl— If you specify the input data as a tableTbl, thenpredictionscan specify the name of one or more variables inTbl, where each variable contains the predicted class labels for one model. In this case, you must specifypredictionsas a character vector, string array, or cell array of character vectors. For example, if the table contains one vector of class labels, stored asTbl.Pred, then specifypredictionsas"Pred".Matrix — Each row of the matrix corresponds to a sample, and each column corresponds to one model's predicted class labels. The number of rows in
predictionsmust be equal to the number of samples inYorTbl. The predicted class labels inpredictionsmust be elements of the true class label variable, andpredictionsmust have the same data type as the true class label variable.
Note
If you specify predicted labels for one or more binary classification models,
fairnessMetrics computes fairness metrics for each model that
returned predicted labels.
Example: Predictions="Pred"
Example: Predictions=["SVMPred","TreePred"]
Data Types: single | double | logical | char | string | cell | categorical
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: Predictions="P",Weights="W" specifies the variables
P and W in the table Tbl as
the model predictions and observation weights, respectively.
Since R2023a
Names of the models with the predicted class labels predictions, specified as a character vector, string array of unique
names, or cell array of unique character vectors.
When
predictionsis an array of names inTbl, the order of the names inModelNamesmust correspond to the order of the names inpredictions. That is,ModelNames{1}is the name of the model with predicted labels in thepredictions{1}table variable,ModelNames{2}is the name of the model with predicted labels in thepredictions{2}table variable, and so on.numel(ModelNames)andnumel(predictions)must be equal. By default, theModelNamesvalue is equivalent topredictions.When
predictionsis a matrix, the order of the names inModelNamesmust correspond to the column order ofpredictions. That is,ModelNames{1}is the name of the model with the predicted labelspredictions(:,1),ModelNames{2}is the name of the model with the predicted labelspredictions(:,2), and so on.numel(ModelNames)andsize(predictions,2)must be equal. By default, theModelNamesvalue is{'Model1','Model2',...}.When
predictionsis[], theModelNamesvalue is'Model1'.Note
You cannot specify the
ModelNamesvalue when thepredictionsvalue is[].
Example: ModelNames="Ensemble"
Example: ModelNames=["SVM","Tree"]
Data Types: char | string | cell
Label of the positive class, specified as a scalar.
PositiveClass must have the same data type as the true class
label variable.
The default PositiveClass value is the second class of the
binary labels, according to the order returned by the unique function with the "sorted" option specified
for the true class label variable.
Example: PositiveClass=categorical(">50K")
Data Types: categorical | char | string | logical | single | double | cell
Reference group for each sensitive attribute, specified as a numeric vector,
string array, or cell array. Each element in the ReferenceGroup
value must have the same data type as the corresponding sensitive attribute. If the
sensitive attributes have mixed types, specify ReferenceGroup
as a cell array. The number of elements in the ReferenceGroup
value must match the number of sensitive attributes.
The default ReferenceGroup value is a vector containing the
mode of each sensitive attribute. The mode is the most frequently occurring value
without taking into account observation weights.
Example: ReferenceGroup={30,categorical("Married-civ-spouse")}
Data Types: single | double | string | cell
Observation weights, specified as a vector of scalar values or the name of a
variable in Tbl. The
software weights the observations in each row of SensitiveAttributes or Tbl with the corresponding
value in Weights. The size of Weights must
equal the number of rows in SensitiveAttributes or
Tbl.
If you specify the input data as a table Tbl, then
Weights can be the name of a variable in
Tbl that contains a numeric vector. In this case, you must
specify Weights as a character vector or string scalar. For
example, if the weights vector W is stored in
Tbl.W, then specify Weights as
"W".
Example: Weights="W"
Data Types: single | double | char | string
Properties
This property is read-only.
Bias metrics, specified as a table.
fairnessMetrics computes the bias metrics for each group in each
sensitive attribute, compared to the reference group of the attribute.
Each row of BiasMetrics contains the bias metrics for a group
in a sensitive attribute.
For data-level evaluation, the first and second variables in
BiasMetricscorrespond to the sensitive attribute name (SensitiveAttributeNamescolumn) and the group name (Groupscolumn), respectively.For model-level evaluation, the first variable corresponds to the model name (
ModelNamescolumn). The second and third variables correspond to the sensitive attribute name and the group name, respectively.
The rest of the variables correspond to the bias metrics in this table.
| Metric Name | Description | Evaluation Type |
|---|---|---|
StatisticalParityDifference | Statistical parity difference (SPD) | Data-level or model-level evaluation |
DisparateImpact | Disparate impact (DI) | Data-level or model-level evaluation |
EqualOpportunityDifference | Equal opportunity difference (EOD) | Model-level evaluation |
AverageAbsoluteOddsDifference | Average absolute odds difference (AAOD) | Model-level evaluation |
The supported bias metrics depend on whether you specify predicted labels by using
the Predictions
argument when you create a fairnessMetrics object.
Data-level evaluation — If you specify true labels and do not specify predicted labels, the
BiasMetricsproperty contains onlyStatisticalParityDifferenceandDisparateImpact.Model-level evaluation — If you specify both true labels and predicted labels, the
BiasMetricsproperty contains all metrics listed in the table.
For definitions of the bias metrics, see Bias Metrics.
Data Types: table
This property is read-only.
Group metrics, specified as a table.
The fairnessMetrics function computes the group metrics for each
group in each sensitive attribute. Note that the function does not use the observation
weights (specified by the Weights
name-value argument) to count the number of samples in each group
(GroupCount value). The function uses Weights
to compute the other metrics.
Each row of GroupMetrics contains the group metrics for a group
in a sensitive attribute.
For data-level evaluation, the first and second variables in
GroupMetricscorrespond to the sensitive attribute name (SensitiveAttributeNamescolumn) and the group name (Groupscolumn), respectively.For model-level evaluation, the first variable corresponds to the model name (
ModelNamescolumn). The second and third variables correspond to the sensitive attribute name and the group name, respectively.
The rest of the variables correspond to the group metrics in this table.
| Metric Name | Description | Evaluation Type |
|---|---|---|
GroupCount | Group count, or number of samples in the group | Data-level or model-level evaluation |
GroupSizeRatio | Group count divided by the total number of samples | Data-level or model-level evaluation |
TruePositives | Number of true positives (TP) | Model-level evaluation |
TrueNegatives | Number of true negatives (TN) | Model-level evaluation |
FalsePositives | Number of false positives (FP) | Model-level evaluation |
FalseNegatives | Number of false negatives (FN) | Model-level evaluation |
TruePositiveRate | True positive rate (TPR), also known as recall or sensitivity,
TP/(TP+FN) | Model-level evaluation |
TrueNegativeRate | True negative rate (TNR), or specificity,
TN/(TN+FP) | Model-level evaluation |
FalsePositiveRate | False positive rate (FPR), also known as fallout or 1-specificity,
FP/(TN+FP) | Model-level evaluation |
FalseNegativeRate | False negative rate (FNR), or miss rate,
FN/(TP+FN) | Model-level evaluation |
FalseDiscoveryRate | False discovery rate (FDR), FP/(TP+FP) | Model-level evaluation |
FalseOmissionRate | False omission rate (FOR), FN/(TN+FN) | Model-level evaluation |
PositivePredictiveValue | Positive predictive value (PPV), or precision,
TP/(TP+FP) | Model-level evaluation |
NegativePredictiveValue | Negative predictive value (NPV), TN/(TN+FN) | Model-level evaluation |
RateOfPositivePredictions | Rate of positive predictions (RPP),
(TP+FP)/(TP+FN+FP+TN) | Model-level evaluation |
RateOfNegativePredictions | Rate of negative predictions (RNP),
(TN+FN)/(TP+FN+FP+TN) | Model-level evaluation |
Accuracy | Accuracy, (TP+TN)/(TP+FN+FP+TN) | Model-level evaluation |
The supported group metrics depend on whether you specify predicted labels by using
the Predictions
argument when you create a fairnessMetrics object.
Data-level evaluation — If you specify true labels and do not specify predicted labels, the
GroupMetricsproperty contains onlyGroupCountandGroupSizeRatio.Model-level evaluation — If you specify both true labels and predicted labels, the
GroupMetricsproperty contains all metrics listed in the table.
Data Types: table
Since R2023a
This property is read-only.
Names of the models with the predicted class labels predictions,
specified as a character vector or cell array of unique character vectors. (The software treats string arrays as cell arrays of character
vectors.)
The ModelNames
name-value argument sets this property.
Data Types: char | cell
This property is read-only.
Label of the positive class, specified as a scalar. (The software treats a string scalar as a character vector.)
The PositiveClass
name-value argument sets this property.
Data Types: categorical | char | logical | single | double | cell
This property is read-only.
Reference group, specified as a numeric vector or cell array. (The software treats string arrays as cell arrays of character vectors.)
The ReferenceGroup name-value argument sets this property.
Data Types: single | double | cell
This property is read-only.
Name of the true class label variable, specified as a character vector containing the name of the response variable. (The software treats a string scalar as a character vector.)
If you specify the
ResponseNameargument, then the specified value determines this property.If you specify
Y, then the property value is'Y'.
Data Types: char
This property is read-only.
Names of the sensitive attribute variables, specified as a character vector or cell array of unique character vectors. (The software treats string arrays as cell arrays of character vectors.)
The sensitiveAttributeNames argument sets this property.
Data Types: char | cell
Examples
Compute fairness metrics for true labels with respect to sensitive attributes by creating a fairnessMetrics object. Then, create a table of fairness metrics by using the report function, and plot bar graphs of the metrics by using the plot function.
Load the sample data census1994, which contains the training data adultdata and the test data adulttest. The data sets consist of demographic information from the US Census Bureau that can be used to predict whether an individual makes over $50,000 per year. Preview the first few rows of the training data set.
load census1994
head(adultdata) age workClass fnlwgt education education_num marital_status occupation relationship race sex capital_gain capital_loss hours_per_week native_country salary
___ ________________ __________ _________ _____________ _____________________ _________________ _____________ _____ ______ ____________ ____________ ______________ ______________ ______
39 State-gov 77516 Bachelors 13 Never-married Adm-clerical Not-in-family White Male 2174 0 40 United-States <=50K
50 Self-emp-not-inc 83311 Bachelors 13 Married-civ-spouse Exec-managerial Husband White Male 0 0 13 United-States <=50K
38 Private 2.1565e+05 HS-grad 9 Divorced Handlers-cleaners Not-in-family White Male 0 0 40 United-States <=50K
53 Private 2.3472e+05 11th 7 Married-civ-spouse Handlers-cleaners Husband Black Male 0 0 40 United-States <=50K
28 Private 3.3841e+05 Bachelors 13 Married-civ-spouse Prof-specialty Wife Black Female 0 0 40 Cuba <=50K
37 Private 2.8458e+05 Masters 14 Married-civ-spouse Exec-managerial Wife White Female 0 0 40 United-States <=50K
49 Private 1.6019e+05 9th 5 Married-spouse-absent Other-service Not-in-family Black Female 0 0 16 Jamaica <=50K
52 Self-emp-not-inc 2.0964e+05 HS-grad 9 Married-civ-spouse Exec-managerial Husband White Male 0 0 45 United-States >50K
Each row contains the demographic information for one adult. The information includes sensitive attributes, such as age, marital_status, relationship, race, and sex. The third column flnwgt contains observation weights, and the last column salary shows whether a person has a salary less than or equal to $50,000 per year (<=50K) or greater than $50,000 per year (>50K).
This example evaluates the fairness of the salary variable with respect to age. Group the age variable into four bins.
ageGroups = ["Age<30","30<=Age<45","45<=Age<60","Age>=60"]; adultdata.age_group = discretize(adultdata.age, ... [min(adultdata.age) 30 45 60 max(adultdata.age)], ... categorical=ageGroups);
Plot the counts of individuals in each class (<=50K and >50K) by age.
figure gc = groupcounts(adultdata,["age_group","salary"]); bar([gc.GroupCount(1:2:end),gc.GroupCount(2:2:end)]) xticklabels(ageGroups) xlabel("Age Group") ylabel("Group Count") legend(["<=50K",">50K"]) grid on
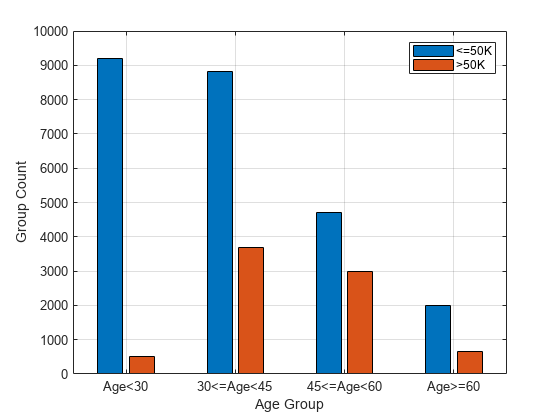
Compute fairness metrics for the salary variable with respect to the age_group variable by using fairnessMetrics.
metricsResults = fairnessMetrics(adultdata,"salary", ... SensitiveAttributeNames="age_group",Weights="fnlwgt")
metricsResults =
fairnessMetrics with properties:
SensitiveAttributeNames: 'age_group'
ReferenceGroup: '30<=Age<45'
ResponseName: 'salary'
PositiveClass: >50K
BiasMetrics: [4×4 table]
GroupMetrics: [4×4 table]
Properties, Methods
metricsResults is a fairnessMetrics object. By default, the fairnessMetrics function selects the majority group of the sensitive attribute (group with the largest number of individuals) as the reference group for the attribute. Also, the fairnessMetrics function orders the labels by using the unique function with the "sorted" option, and specifies the second class of the labels as the positive class. In this data set, the reference group of age_group is the group 30<=Age<45, and the positive class is >50K. The object stores bias metrics and group metrics in the BiasMetrics and GroupMetrics properties, respectively. Display the properties.
metricsResults.BiasMetrics
ans=4×4 table
SensitiveAttributeNames Groups StatisticalParityDifference DisparateImpact
_______________________ __________ ___________________________ _______________
age_group Age<30 -0.24365 0.17661
age_group 30<=Age<45 0 1
age_group 45<=Age<60 0.098497 1.3329
age_group Age>=60 -0.05041 0.82965
metricsResults.GroupMetrics
ans=4×4 table
SensitiveAttributeNames Groups GroupCount GroupSizeRatio
_______________________ __________ __________ ______________
age_group Age<30 9711 0.29824
age_group 30<=Age<45 12489 0.38356
age_group 45<=Age<60 7717 0.237
age_group Age>=60 2644 0.081201
According to the bias metrics, the salary variable is biased toward the age group 45 to 60 years and biased against the age group less than 30 years, compared to the reference group (30<=Age<45).
You can create a table that contains both bias metrics and group metrics by using the report function. Specify GroupMetrics as "all" to include all group metrics. You do not have to specify the BiasMetrics name-value argument because its default value is "all".
metricsTbl = report(metricsResults,GroupMetrics="all")metricsTbl=4×6 table
SensitiveAttributeNames Groups StatisticalParityDifference DisparateImpact GroupCount GroupSizeRatio
_______________________ __________ ___________________________ _______________ __________ ______________
age_group Age<30 -0.24365 0.17661 9711 0.29824
age_group 30<=Age<45 0 1 12489 0.38356
age_group 45<=Age<60 0.098497 1.3329 7717 0.237
age_group Age>=60 -0.05041 0.82965 2644 0.081201
Visualize the bias metrics by using the plot function.
figure t = tiledlayout(2,1); nexttile plot(metricsResults,"spd") xlabel("") ylabel("") nexttile plot(metricsResults,"di") xlabel("") ylabel("") xlabel(t,"Fairness Metric Value") ylabel(t,"Age Group")
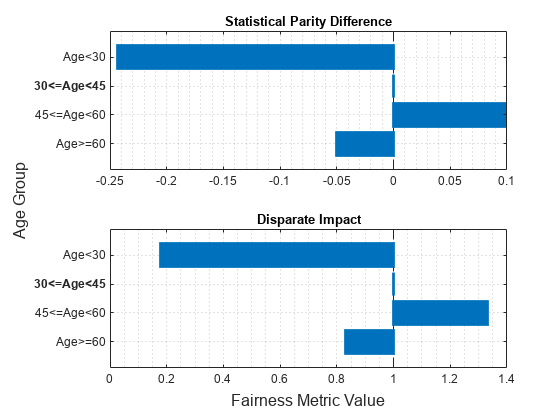
The vertical line in each plot ( for statistical parity difference and for disparate impact) indicates the metric value for the reference group. If the labels do not have a bias for a target group compared to the reference group, the metric value for the target group is the same as the metric value for the reference group.
Compute fairness metrics for predicted labels with respect to sensitive attributes by creating a fairnessMetrics object. Then, create a table of fairness metrics by using the report function, and plot bar graphs of the metrics by using the plot function.
Load the sample data census1994, which contains the training data adultdata and the test data adulttest. The data sets consist of demographic information from the US Census Bureau that can be used to predict whether an individual makes over $50,000 per year. Preview the first few rows of the training data set.
load census1994
head(adultdata) age workClass fnlwgt education education_num marital_status occupation relationship race sex capital_gain capital_loss hours_per_week native_country salary
___ ________________ __________ _________ _____________ _____________________ _________________ _____________ _____ ______ ____________ ____________ ______________ ______________ ______
39 State-gov 77516 Bachelors 13 Never-married Adm-clerical Not-in-family White Male 2174 0 40 United-States <=50K
50 Self-emp-not-inc 83311 Bachelors 13 Married-civ-spouse Exec-managerial Husband White Male 0 0 13 United-States <=50K
38 Private 2.1565e+05 HS-grad 9 Divorced Handlers-cleaners Not-in-family White Male 0 0 40 United-States <=50K
53 Private 2.3472e+05 11th 7 Married-civ-spouse Handlers-cleaners Husband Black Male 0 0 40 United-States <=50K
28 Private 3.3841e+05 Bachelors 13 Married-civ-spouse Prof-specialty Wife Black Female 0 0 40 Cuba <=50K
37 Private 2.8458e+05 Masters 14 Married-civ-spouse Exec-managerial Wife White Female 0 0 40 United-States <=50K
49 Private 1.6019e+05 9th 5 Married-spouse-absent Other-service Not-in-family Black Female 0 0 16 Jamaica <=50K
52 Self-emp-not-inc 2.0964e+05 HS-grad 9 Married-civ-spouse Exec-managerial Husband White Male 0 0 45 United-States >50K
Each row contains the demographic information for one adult. The information includes sensitive attributes, such as age, marital_status, relationship, race, and sex. The third column flnwgt contains observation weights, and the last column salary shows whether a person has a salary less than or equal to $50,000 per year (<=50K) or greater than $50,000 per year (>50K).
Train a classification tree using the training data set adultdata. Specify the response variable, predictor variables, and observation weights by using the variable names in the adultdata table.
predictorNames = ["capital_gain","capital_loss","education", ... "education_num","hours_per_week","occupation","workClass"]; Mdl = fitctree(adultdata,"salary", ... PredictorNames=predictorNames,Weights="fnlwgt");
Predict the test sample labels by using the trained tree Mdl.
labels = predict(Mdl,adulttest);
This example evaluates the fairness of the predicted labels with respect to age and marital status. Group the age variable into four bins.
ageGroups = ["Age<30","30<=Age<45","45<=Age<60","Age>=60"]; adulttest.age_group = discretize(adulttest.age, ... [min(adulttest.age) 30 45 60 max(adulttest.age)], ... categorical=ageGroups);
Plot the counts of individuals in each predicted class (<=50K and >50K) by age.
figure
gs_age = groupcounts({adulttest.age_group,labels});
b_age = bar([gs_age(1:2:end),gs_age(2:2:end)]);
xticklabels(ageGroups)
xlabel("Age Group")
ylabel("Group Count")
legend(["<=50K",">50K"])
grid minor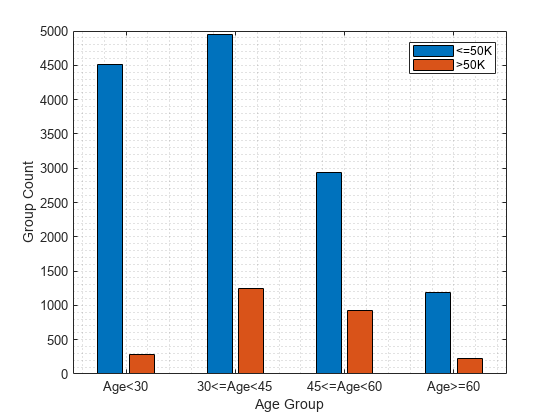
Plot the counts of individuals by marital status. Display the count values near the tips of the bars if the values are smaller than 100.
figure
gs_status = groupcounts({adulttest.marital_status,labels});
b_status = bar([gs_status(1:2:end),gs_status(2:2:end)]);
xticklabels(unique(adulttest.marital_status))
xlabel("Marital Status")
ylabel("Group Count")
legend(["<=50K",">50K"])
grid minor
xtips1 = b_status(1).XEndPoints;
ytips1 = b_status(1).YEndPoints;
labels1 = string(b_status(1).YData);
ind1 = ytips1 < 100;
text(xtips1(ind1),ytips1(ind1),labels1(ind1), ...
HorizontalAlignment="center",VerticalAlignment="bottom", ...
Color=b_status(1).FaceColor)
xtips2 = b_status(2).XEndPoints;
ytips2 = b_status(2).YEndPoints;
labels2 = string(b_status(2).YData);
ind2 = ytips2 < 100;
text(xtips2(ind2),ytips2(ind2),labels2(ind2), ...
HorizontalAlignment="center",VerticalAlignment="bottom", ...
Color=b_status(2).FaceColor)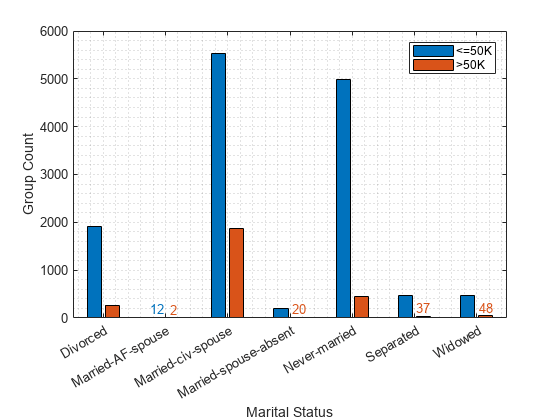
Compute fairness metrics for the predictions (labels) with respect to the age_group and marital_status variables by using fairnessMetrics.
metricsResults = fairnessMetrics(adulttest,"salary", ... SensitiveAttributeNames=["age_group","marital_status"], ... Predictions=labels,Weights="fnlwgt")
metricsResults =
fairnessMetrics with properties:
SensitiveAttributeNames: {'age_group' 'marital_status'}
ReferenceGroup: {'30<=Age<45' 'Married-civ-spouse'}
ResponseName: 'salary'
PositiveClass: >50K
BiasMetrics: [11×7 table]
GroupMetrics: [11×20 table]
ModelNames: 'Model1'
Properties, Methods
metricsResults is a fairnessMetrics object. By default, the fairnessMetrics function selects the majority group of each sensitive attribute (group with the largest number of individuals) as the reference group for the attribute. Also, the fairnessMetrics function orders the labels by using the unique function with the "sorted" option, and specifies the second class of the labels as the positive class. In this data set, the reference groups of age_group and marital_status are the groups 30<=Age<45 and Married-civ-spouse, respectively, and the positive class is >50K. The object stores bias metrics and group metrics in the BiasMetrics and GroupMetrics properties, respectively.
Create a table with fairness metrics by using the report function. Specify BiasMetrics as ["eod","aaod"] to include the equal opportunity difference (EOD) and average absolute odds difference (AAOD) metrics in the report table. The fairnessMetrics function computes the two metrics by using the true positive rates (TPR) and false positive rates (FPR). Specify GroupMetrics as ["tpr","fpr"] to include TPR and FPR values in the table.
metricsTbl = report(metricsResults, ... BiasMetrics=["eod","aaod"],GroupMetrics=["tpr","fpr"])
metricsTbl=11×7 table
ModelNames SensitiveAttributeNames Groups EqualOpportunityDifference AverageAbsoluteOddsDifference TruePositiveRate FalsePositiveRate
__________ _______________________ _____________________ __________________________ _____________________________ ________________ _________________
Model1 age_group Age<30 -0.041319 0.044114 0.41333 0.041709
Model1 age_group 30<=Age<45 0 0 0.45465 0.088618
Model1 age_group 45<=Age<60 0.061495 0.031809 0.51614 0.086495
Model1 age_group Age>=60 0.0060387 0.011955 0.46069 0.070746
Model1 marital_status Divorced 0.078541 0.043643 0.54263 0.075653
Model1 marital_status Married-AF-spouse 0.073166 0.078782 0.53726 0
Model1 marital_status Married-civ-spouse 0 0 0.46409 0.084398
Model1 marital_status Married-spouse-absent -0.067098 0.048093 0.39699 0.055311
Model1 marital_status Never-married 0.0886 0.057557 0.55269 0.057883
Model1 marital_status Separated 0.027256 0.026751 0.49135 0.058151
Model1 marital_status Widowed 0.12442 0.080073 0.58851 0.048675
Plot the EOD and AAOD values for the sensitive attribute age_group. Because age_group is the first element in the SensitiveAttributeNames property of metricsResults, it is the default value for the property. Therefore, you do not have to specify the SensitiveAttributeName argument of the plot function.
figure t = tiledlayout(1,2); nexttile plot(metricsResults,"eod") title("EOD") xlabel("") ylabel("") nexttile plot(metricsResults,"aaod") title("AAOD") xlabel("") ylabel("") yticklabels("") xlabel(t,"Fairness Metric Value") ylabel(t,"Age Group")
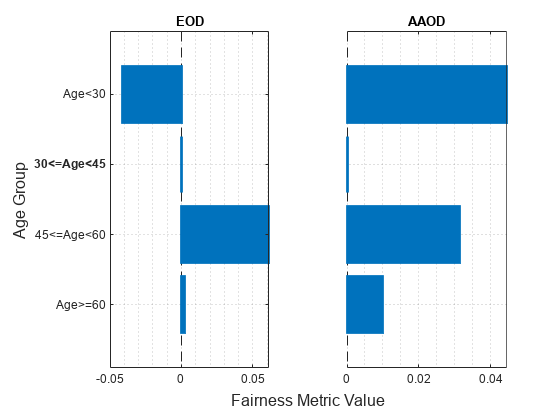
The vertical line at indicates the metric value for the reference group (30<=Age<45). If the labels do not have a bias for a target group compared to the reference group, the metric value for the target group is the same as the metric value for the reference group. According to the EOD values (differences in TPR), the predictions for the salary variable are most biased toward the group 45<=Age<60 compared to the reference group. According to the AAOD values (averaged differences in TPR and FPR), the predictions are most biased toward the group Age<30.
Plot the EOD and AAOD values for the sensitive attribute marital_status by specifying the SensitiveAttributeName argument of the plot function as marital_status.
figure t = tiledlayout(1,2); nexttile plot(metricsResults,"eod",SensitiveAttributeName="marital_status") title("EOD") xlabel("") ylabel("") nexttile plot(metricsResults,"aaod",SensitiveAttributeName="marital_status") title("AAOD") xlabel("") ylabel("") yticklabels("") xlabel(t,"Fairness Metric Value") ylabel(t,"Marital Status")
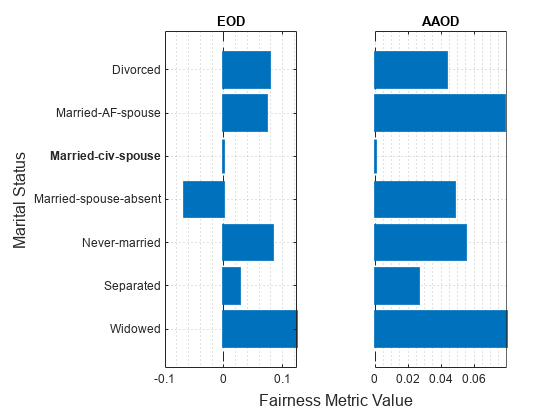
The vertical line at indicates the metric value for the reference group (Married-civ-spouse). According to the EOD values, the predictions for the salary variable are most biased toward the group Widowed compared to the reference group. According to the AAOD values, the predictions are similarly biased toward the groups Widowed and Married-AF-spouse.
Train two classification models, and compare the model predictions by using fairness metrics.
Read the sample file CreditRating_Historical.dat into a table. The predictor data consists of financial ratios and industry sector information for a list of corporate customers. The response variable consists of credit ratings assigned by a rating agency.
creditrating = readtable("CreditRating_Historical.dat");Because each value in the ID variable is a unique customer ID—that is, length(unique(creditrating.ID)) is equal to the number of observations in creditrating—the ID variable is a poor predictor. Remove the ID variable from the table, and convert the Industry variable to a categorical variable.
creditrating.ID = []; creditrating.Industry = categorical(creditrating.Industry);
In the Rating response variable, combine the AAA, AA, A, and BBB ratings into a category of "good" ratings, and the BB, B, and CCC ratings into a category of "poor" ratings.
Rating = categorical(creditrating.Rating); Rating = mergecats(Rating,["AAA","AA","A","BBB"],"good"); Rating = mergecats(Rating,["BB","B","CCC"],"poor"); creditrating.Rating = Rating;
Train a support vector machine (SVM) model on the creditrating data. For better results, standardize the predictors before fitting the model. Use the trained model to predict labels and compute the misclassification rate for the training data set.
predictorNames = ["WC_TA","RE_TA","EBIT_TA","MVE_BVTD","S_TA"]; SVMMdl = fitcsvm(creditrating,"Rating", ... PredictorNames=predictorNames,Standardize=true); SVMPredictions = resubPredict(SVMMdl); resubLoss(SVMMdl)
ans = 0.0872
Train a generalized additive model (GAM).
GAMMdl = fitcgam(creditrating,"Rating", ... PredictorNames=predictorNames); GAMPredictions = resubPredict(GAMMdl); resubLoss(GAMMdl)
ans = 0.0542
GAMMdl achieves better accuracy on the training data set.
Compute fairness metrics with respect to the sensitive attribute Industry by using the model predictions for both models.
predictions = [SVMPredictions,GAMPredictions]; metricsResults = fairnessMetrics(creditrating,"Rating", ... SensitiveAttributeNames="Industry",Predictions=predictions, ... ModelNames=["SVM","GAM"]);
Display the bias metrics by using the report function.
report(metricsResults)
ans=48×5 table
Metrics SensitiveAttributeNames Groups SVM GAM
___________________________ _______________________ ______ _________ __________
StatisticalParityDifference Industry 1 -0.028441 0.0058208
StatisticalParityDifference Industry 2 -0.04014 0.0063339
StatisticalParityDifference Industry 3 0 0
StatisticalParityDifference Industry 4 -0.04905 -0.0043007
StatisticalParityDifference Industry 5 -0.015615 0.0041607
StatisticalParityDifference Industry 6 -0.03818 -0.024515
StatisticalParityDifference Industry 7 -0.01514 0.007326
StatisticalParityDifference Industry 8 0.0078632 0.036581
StatisticalParityDifference Industry 9 -0.013863 0.042266
StatisticalParityDifference Industry 10 0.0090218 0.050095
StatisticalParityDifference Industry 11 -0.004188 0.001453
StatisticalParityDifference Industry 12 -0.041572 -0.028589
DisparateImpact Industry 1 0.92261 1.017
DisparateImpact Industry 2 0.89078 1.0185
DisparateImpact Industry 3 1 1
DisparateImpact Industry 4 0.86654 0.98742
⋮
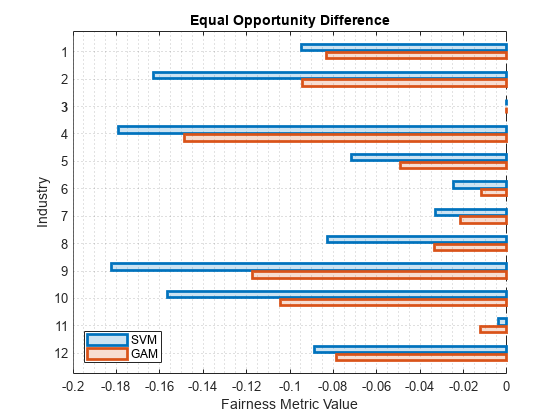
Among the bias metrics, compare the equal opportunity difference (EOD) values. Create a bar graph of the EOD values by using the plot function.
b = plot(metricsResults,"eod"); b(1).FaceAlpha = 0.2; b(2).FaceAlpha = 0.2; legend(Location="southwest")
To better understand the distributions of EOD values, plot the values using box plots.
boxchart(metricsResults.BiasMetrics.EqualOpportunityDifference, ... GroupByColor=metricsResults.BiasMetrics.ModelNames) ax = gca; ax.XTick = []; ylabel("Equal Opportunity Difference") legend
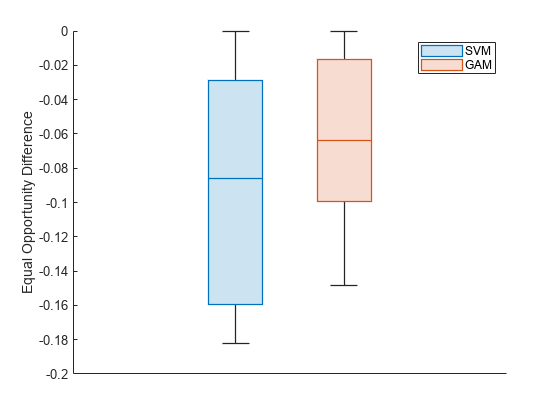
The EOD values for GAM are closer to 0 compared to the values for SVM.
More About
The fairnessMetrics object supports four bias metrics:
statistical parity difference (SPD), disparate impact (DI), equal opportunity difference
(EOD), and average absolute odds difference (AAOD). The object supports EOD and AAOD only
for evaluating model predictions.
A fairnessMetrics object computes bias metrics for each group in each
sensitive attribute with respect to the reference group of the attribute.
Statistical parity (or demographic parity) difference (SPD)
The SPD value of the ith sensitive attribute (Si) for the group sij with respect to the reference group sir is defined by
The SPD value is the difference between the probability of being in the positive class when the sensitive attribute value is sij and the probability of being in the positive class when the sensitive attribute value is sir (reference group). This metric assumes that the two probabilities (statistical parities) are equal if the labels are unbiased with respect to the sensitive attribute.
If you specify the
Predictionsargument, the software computes SPD for the probabilities of the model predictions instead of the true labels Y.Disparate impact (DI)
The DI value of the ith sensitive attribute (Si) for the group sij with respect to the reference group sir is defined by
The DI value is the ratio of the probability of being in the positive class when the sensitive attribute value is sij to the probability of being in the positive class when the sensitive attribute value is sir (reference group). This metric assumes that the two probabilities are equal if the labels are unbiased with respect to the sensitive attribute. In general, a DI value less than
0.8or greater than1.25indicates bias with respect to the reference group [2].If you specify the
Predictionsargument, the software computes DI for the probabilities of the model predictions instead of the true labels Y.Equal opportunity difference (EOD)
The EOD value of the ith sensitive attribute (Si) for the group sij with respect to the reference group sir is defined by
The EOD value is the difference in the true positive rate (TPR) between the group sij and the reference group sir. This metric assumes that the two rates are equal if the predicted labels are unbiased with respect to the sensitive attribute.
Average absolute odds difference (AAOD)
The AAOD value of the ith sensitive attribute (Si) for the group sij with respect to the reference group sir is defined by
The AAOD value represents the difference in the true positive rates (TPR) and false positive rates (FPR) between the group sij and the reference group sir. This metric assumes no difference in TPR and FPR if the predicted labels are unbiased with respect to the sensitive attribute.
Algorithms
fairnessMetrics considers NaN, ''
(empty character vector), "" (empty string),
<missing>, and <undefined> values in
Tbl, Y, and
SensitiveAttributes to be missing values.
fairnessMetrics does not use observations with missing values.
References
[1] Mehrabi, Ninareh, et al. “A Survey on Bias and Fairness in Machine Learning.” ArXiv:1908.09635 [cs.LG], Sept. 2019. arXiv.org.
[2] Saleiro, Pedro, et al. “Aequitas: A Bias and Fairness Audit Toolkit.” ArXiv:1811.05577 [cs.LG], April 2019. arXiv.org.
Version History
Introduced in R2022bYou can compare fairness metrics across multiple binary classifiers by using the fairnessMetrics
function. In the call to the function, use the predictions argument and
specify the predicted class labels for each model. To specify the names of the models, you
can use the ModelNames name-value argument. The model name information
is stored in the BiasMetrics, GroupMetrics, and
ModelNames properties of the fairnessMetrics
object.
After you create a fairnessMetrics object, use the report or plot object function.
The
reportobject function returns a fairness metrics table, whose format depends on the value of theDisplayMetricsInRowsname-value argument. (For more information, seemetricsTbl.) You can specify a subset of models to include in the report table by using theModelNamesname-value argument.The
plotobject function returns a bar graph as an array ofBarobjects. The bar colors indicate the models whose predicted labels are used to compute the specified metric. You can specify a subset of models to include in the plot by using theModelNamesname-value argument.
In previous releases, the first and second variables in the
BiasMetrics and GroupMetrics properties of the
fairnessMetrics object always corresponded to the sensitive attribute
name (SensitiveAttributeNames column) and the group name
(Groups column), respectively. For more information on the current
behavior, see BiasMetrics and
GroupMetrics.
See Also
Topics
- Introduction to Fairness in Binary Classification
- Explore Fairness Metrics for Credit Scoring Model (Risk Management Toolbox)
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)