Choose Between spmd, parfor, and
parfeval
To run computations in parallel, you can use the spmd, parfor, parfeval, or parfevalOnAll functions. Each function
relies on different parallel programming concepts.
When to Use parfor
A parfor-loop can be useful if you have a
for-loop with slow loop iterations or many loop iterations.
Each iteration must be independent of all others. For more help in deciding when to
use parfor, see Decide When to Use parfor.
When to Use spmd
Run Communicating Parallel Code
Use spmd if you require fine-grained worker-to-worker
communication and collaboration between workers during a computation.
parfor, parfeval, and
parfevalOnAll do not allow communication between
workers. Computations with spmd can involve communication
between workers using the spmdSend, spmdReceive, and spmdSendReceive functions.
If you are unsure, ask yourself the following: within my parallel
code, can each computation be completed without any communication between
workers? If yes, use parfor or
parfeval. Otherwise, use
spmd.
Run Parallel and Customized Code on Distributed Arrays
Use spmd if your computations involve large arrays
distributed across workers. You can perform simultaneous calculations on all
workers or perform customized calculations on specific workers.
When workers run an spmd block, each worker is assigned a
unique index, the spmdIndex. This lets you specify code to be run only on certain
workers, and target sections of distributed arrays.
Synchronous and Asynchronous Work
When choosing between parfor,
parfeval, and spmd, consider
whether your calculation requires synchronization with the client.
parfor and spmd require
synchronization, and therefore block you from running any new computations on
the MATLAB® client. parfeval does not require
synchronization, so you can continue to use the client.
Use Other Functional Capabilities
spmd and parfeval have other
capabilities you can use after you have submitted your computations.
With
spmd, you can collect results computed inside thespmdstatement without transferring the results to the client. Access the values of the variables assigned inside thespmdstatement asCompositeobjects from the client. For more information, see Access Worker Variables with Composites.When you submit a
parfevaltask, MATLAB schedules the task to run asynchronously and returnsFutureobjects before the submitted task finishes running. AFutureobject represents the task that MATLAB has scheduled. You can interact withFutureobjects in different ways:
Compare Performance of parfor, parfeval, and spmd
Using spmd can be slower or faster than using parfor-loops or parfeval, depending on the type of computation. Overhead affects the relative performance of parfor-loops, parfeval, and spmd.
For a set of tasks, parfor and parfeval typically perform better than spmd under these conditions:
The computational time taken per task is not deterministic.
The computational time taken per task is not uniform.
The data returned from each task is small.
Use parfeval when:
You want to run computations in the background.
Each task is dependent on other tasks.
In this example, you examine the speed at which the software performs matrix operations when using a parfor-loop, parfeval, and spmd.
First, create a parallel pool of process workers p.
p = parpool("Processes");Starting parallel pool (parpool) using the 'Processes' profile ... Connected to parallel pool with 6 workers.
Compute Random Matrices
Examine the speed at which the software can generate random matrices by using a parfor-loop, parfeval, and spmd. Set the number of trials (n) and the matrix size (for an m-by-m matrix). Increasing the number of trials improves the statistics used in later analysis, but does not affect the calculation itself.
m =1500; n =
20;
Then, use a parfor-loop to execute rand(m) once for each worker. Time each of the n trials.
parforTime = zeros(n,1); for i = 1:n tic; mats = cell(1,p.NumWorkers); parfor N = 1:p.NumWorkers mats{N} = rand(m); end parforTime(i) = toc; end
Next, use parfeval to execute rand(m) once for each worker. Time each of the n trials.
parfevalTime = zeros(n,1); for i = 1:n tic; f(1:p.NumWorkers) = parallel.FevalFuture; for N = 1:p.NumWorkers f(N) = parfeval(@rand,1,m); end mats = fetchOutputs(f); parfevalTime(i) = toc; clear f end
Finally, use spmd to execute rand(m) once for each worker. You can use spmdCat to concatenate the values of mat on each worker into array mats and store it on worker 1. For details on workers and how to execute commands on them with spmd, see Run Single Programs on Multiple Data Sets. Time each of the n trials.
spmdTime = zeros(n,1); for i = 1:n tic; spmd mat = rand(m); mats = spmdCat({mat}, 1, 1); end allMats = mats{1}; spmdTime(i) = toc; end
Use rmoutliers to remove the outliers from each of the trials. Then, use boxplot to compare the times.
% Hide outliers boxData = rmoutliers([parforTime parfevalTime spmdTime]); % Plot data boxplot(boxData, 'labels',{'parfor','parfeval','spmd'}, 'Symbol','') ylabel('Time (seconds)') title('Make n Random Matrices (m-by-m)')
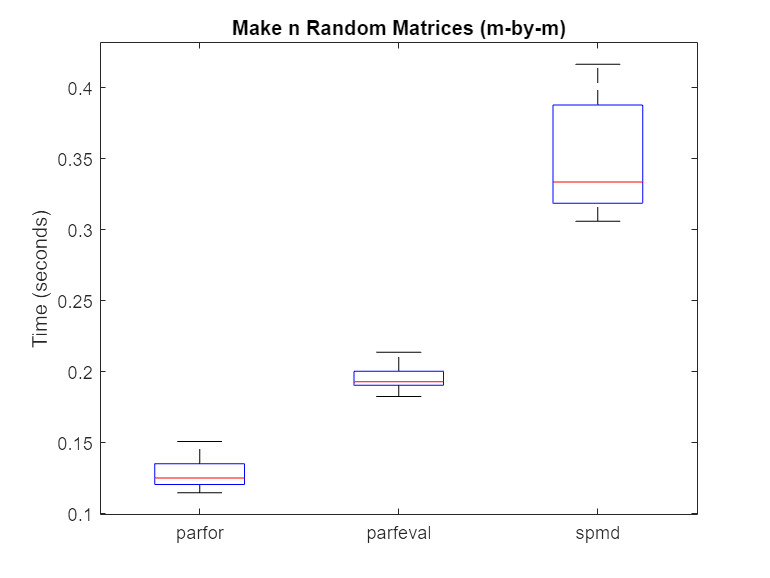
Typically, spmd requires more overhead per evaluation than parfor or parfeval. Therefore, in this case, using a parfor-loop or parfeval is more efficient.
Compute Sum of Random Matrices
Next, compute the sum of random matrices. You can do this by using a reduction variable with a parfor-loop, a sum after computations with parfeval, or spmdPlus with spmd. Again, set the number of trials (n) and the matrix size (for an m-by-m matrix).
m =1500; n =
20;
Then, use a parfor-loop to execute rand(m) once for each worker. Compute the sum with a reduction variable. Time each of the n trials.
parforTime = zeros(n,1); for i = 1:n tic; result = 0; parfor N = 1:p.NumWorkers result = result + rand(m); end parforTime(i) = toc; end
Next, use parfeval to execute rand(m) once for each worker. Use fetchOutputs to fetch all of the matrices, then use sum. Time each of the n trials.
parfevalTime = zeros(n,1); for i = 1:n tic; f(1:p.NumWorkers) = parallel.FevalFuture; for N = 1:p.NumWorkers f(N) = parfeval(@rand,1,m); end result = sum(fetchOutputs(f)); parfevalTime(i) = toc; clear f end
Finally, use spmd to execute rand(m) once for each worker. Use spmdPlus to sum all of the matrices. To send the result only to the first worker, set the optional target worker argument to 1. Time each of the n trials.
spmdTime = zeros(n,1); for i = 1:n tic; spmd r = spmdPlus(rand(m), 1); end result = r{1}; spmdTime(i) = toc; end
Use rmoutliers to remove the outliers from each of the trials. Then, use boxplot to compare the times.
% Hide outliers boxData = rmoutliers([parforTime parfevalTime spmdTime]); % Plot data boxplot(boxData, 'labels',{'parfor','parfeval','spmd'}, 'Symbol','') ylabel('Time (seconds)') title('Sum of n Random Matrices (m-by-m)')
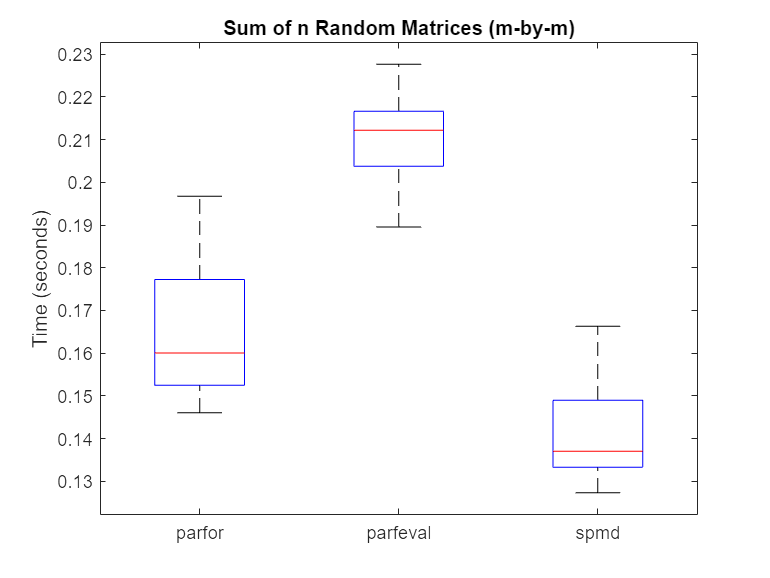
For this calculation, spmd is faster than a parfor-loop or parfeval. When you use reduction variables in a parfor-loop, each worker performs a local reduction before sending its partial result back to the client to compute the final result.
By contrast, spmd calls spmdPlus only once to do a global reduction operation, requiring less overhead. As such, the overhead for the reduction part of the calculation is for spmd, and for parfor.