msdotplot
Plot set of peak lists from LC/MS or GC/MS data set
Syntax
msdotplot(Peaklist, Times)
msdotplot(FigHandle, Peaklist, Times)
msdotplot(..., 'Quantile', QuantileValue)
PlotHandle = msdotplot(...)
Input Arguments
Peaklist | Cell array of peak lists, where each
element is a two-column matrix with m/z values in the first column
and ion intensity values in the second column. Each element corresponds
to a spectrum or retention time. Tip You can use the |
Times | Vector of retention times associated
with an LC/MS or GC/MS data set. The number of elements in Times equals
the number of elements in the cell array Peaklist.Tip You can use the |
FigHandle | Handle to an open Figure window such
as one created by the msheatmap function. |
QuantileValue | Value that specifies a percentage. When
peaks are ranked by intensity, only those that rank above this percentage
are plotted. Choices are any value ≥ 0 and ≤
1. Default is 0. For example, setting QuantileValue = 0 plots
all peaks, and setting QuantileValue = 0.8 plots
only the 20% most intense peaks. |
Output Arguments
PlotHandle | Handle to the line series object (figure plot). |
Description
msdotplot( plots
a set of peak lists from a liquid chromatography/mass spectrometry
(LC/MS) or gas chromatography/mass spectrometry (GC/MS) data set represented
by Peaklist, Times)Peaklist, a cell array of peak lists,
where each element is a two-column matrix with m/z values in the first
column and ion intensity values in the second column, and Times,
a vector of retention times associated with the spectra. Peaklist and Times have
the same number of elements. The data is plotted into any existing
figure generated by the msheatmap function;
otherwise, the data is plotted into a new Figure window.
msdotplot( plots
the set of peak lists into the axes contained in an open Figure window
with the handle FigHandle, Peaklist, Times)FigHandle.
Tip
This syntax is useful to overlay a dot plot on top of a heat
map of mass spectrometry data created with the msheatmap function.
msdotplot(..., 'Quantile', plots
only the most intense peaks, specifically those in the percentage
above the specified QuantileValue)QuantileValue. Choices
are any value ≥ 0 and ≤
1. Default is 0. For example, setting QuantileValue = 0 plots
all peaks, and setting QuantileValue = 0.8 plots
only the 20% most intense peaks.
PlotHandle = msdotplot(...)get function to display
a list of the plot's properties. You can use this handle as input
to the set function to change the plot's properties,
including showing and hiding points.
Examples
Load a MAT-file, included with the Bioinformatics Toolbox™ software, which contains LC/MS data variables, including
peaksandret_time.peaksis a cell array of peak lists, where each element is a two-column matrix of m/z values and ion intensity values, and each element corresponds to a spectrum or retention time.ret_timeis a column vector of retention times associated with the LC/MS data set.load lcmsdataCreate a dot plot with only the 5% most intense peaks.
msdotplot(ms_peaks,ret_time,'Quantile',0.95)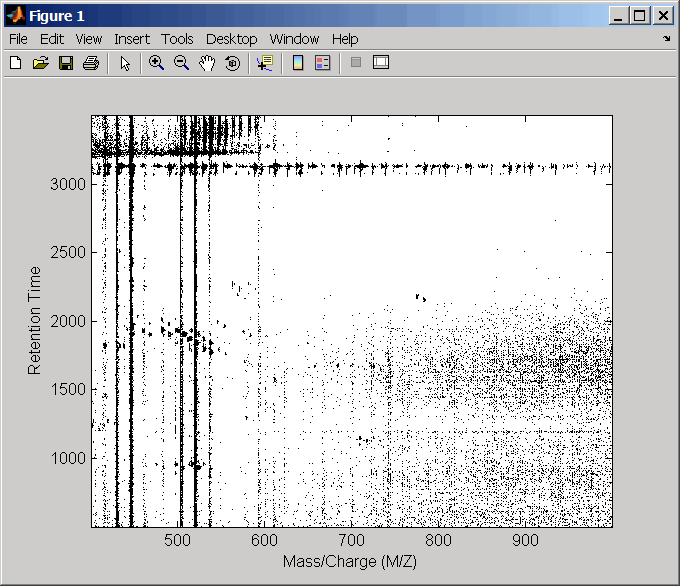
Resample the data, then create a heat map of the LC/MS data.
[MZ,Y] = msppresample(ms_peaks,5000); msheatmap(MZ,ret_time,log(Y))

Overlay the dot plot on the heat map, and then zoom in to see the detail.
msdotplot(ms_peaks,ret_time) axis([480 532 375 485])

Version History
Introduced in R2007a
See Also
mspalign | msbackadj | msalign | msheatmap | mslowess | msnorm | mspeaks | msresample | msppresample | mssgolay | msviewer
Topics
- Mass Spectrometry and Bioanalytics
- Preprocessing Raw Mass Spectrometry Data
- Visualizing and Preprocessing Hyphenated Mass Spectrometry Data Sets for Metabolite and Protein/Peptide Profiling
- Differential Analysis of Complex Protein and Metabolite Mixtures Using Liquid Chromatography/Mass Spectrometry (LC/MS)