Insect Muscle Activation Dynamics Simulator (IMADSim)
Included insect muscle activation dynamic models are from;
Non-linear models:
- Zakotnik et al., 2006
- Wilson et al., 2013
Linear models:
- Zajac, 1989
- Blumel et al., 2012
- Wilson et al., 2012
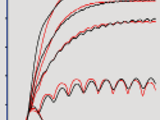
The GUI has five panels to select or set: the model, the corresponding model parameters, the motor neuron type, the spike shape and the spike generator. Arbitrary spike trains can be loaded from a .mat or .txt file consisting of spike times. For constant frequency spike trains, single or multiple spike frequencies can be set. The relaxation time may also be specified. There are three displays showing: the time course of isometric force generation, the corresponding spike train, and the model equations. Hold, Run and Reset buttons are used, respectively, to keep the multiple time courses on the display, run the simulation and to set the GUI to default values. Graphical outputs and the corresponding data can be saved to the workspace by selecting options in the toolbar. Tooltips or hints are provided when the mouse pointer hovers over an item in the GUI.
The optimisation routines are based on the Matlab LSQNONLIN solver for non-linear least squares problems. There are two main routines. The first is for both Zakotnik and Wilson non-linear models. The second is for the Zakotnik model only, and provides intermediate results (see below). When calling functions, several arguments can be set to customize the optimisation. Otherwise, default values will be used. The experimental data should be sampled at 5 kHz.
Experimental data (http://doi.org/10.4119/unibi/2937068) are also provided from six animals (female locusts, Schistocerca gregaria, hind-leg muscle responses for spike trains with different frequencies): three for the Slow Extensor Tibiae motor neuron (SETi) and three for the Fast Extensor Tibiae motor neuron (FETi). Corresponding optimisation results for the two non-linear models are provided.
Zitieren als
Harischandra N., Clare A., Zakotnik J., Blackburn L., Matheson T., and Dürr V. (2019) Evaluation of linear and non-linear activation dynamics models for insect muscles, PLoS Comput Biol, 15(10): e1007437
Clare AJ, Blackburn LML, Harischandra N, Zakotnik J, Dürr V, Matheson T. (2019) Activation dynamics data for locust hind leg extensor muscle, Bielefeld University; (for the experimental data)
Kompatibilität der MATLAB-Version
Plattform-Kompatibilität
Windows macOS LinuxKategorien
- Mathematics and Optimization > Global Optimization Toolbox > Global or Multiple Starting Point Search >
- Sciences > Neuroscience > Neural Simulation >
- Engineering > Mechanical Engineering > Statics and Dynamics >
Tags
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!Live Editor erkunden
Erstellen Sie Skripte mit Code, Ausgabe und formatiertem Text in einem einzigen ausführbaren Dokument.
IMADSim
IMADSim/GUI
IMADSim/Optimisation
| Version | Veröffentlicht | Versionshinweise | |
|---|---|---|---|
| 1.0.4 | Bug fixes |
|
|
| 1.0.3 | Updated README.txt and the IMADSim Documentation file. |
|
|
| 1.0.2 | Add an item to the "Cite As" to represent the experimental data. |
|
|
| 1.0.1 | Add a link to the published DATA in the desceiption. |
|
|
| 1.0.0 |
|