updateMetrics
Update performance metrics in incremental dynamic k-means clustering model given new data
Since R2025a
Description
Mdl = updateMetrics(Mdl,X)Mdl, which is the input
incremental dynamic k-means clustering model Mdl modified to contain the
model performance metrics on the incoming predictor data X.
When the input model is warm (Mdl.IsWarm is true),
updateMetrics overwrites previously computed metrics, stored in the
Metrics and DynamicMetrics properties, with the new
values. Otherwise, updateMetrics stores NaN
values.
Examples
Create a data set with 20,000 observations of three predictors. The data set contains two groups of 10,000 observations each. Store the group identification numbers in ids.
rng(0,"twister"); % For reproducibility ngroups = 2; obspergroup = 10000; Xtrain = []; ids = []; sigma = 0.4; for c = 1:ngroups Xtrain = [Xtrain; randn(obspergroup,3)*sigma + ... (randi(2,[1,3])-1).*ones(obspergroup,3)]; ids = [ids; c*ones(obspergroup,1)]; end
Shuffle the data set.
ntrain = size(Xtrain,1); indices = randperm(ntrain); Xtrain = Xtrain(indices,:); ids = ids(indices,:);
Create a test set that contains the last 2000 observations of the data set. Store the group identification numbers for the test set in idsTest. Keep the first 18,000 observations as the training set.
Xtest = Xtrain(end-1999:end,:); idsTest = ids(end-1999:end,:); Xtrain = Xtrain(1:end-2000,:); ids = ids(1:end-2000,:);
Plot the training set, and color the observations according to their group identification number.
scatter3(Xtrain(:,1),Xtrain(:,2),Xtrain(:,3),1,ids,"filled");
Create Incremental Model
Create an incremental dynamic k-means model object with a warm-up period of 1000 observations. Specify that the incremental fit function stores two clusters that are merged from the dynamic clusters.
Mdl = incrementalDynamicKMeans(numClusters=2, ...
WarmupPeriod=1000, MergeClusters=true)Mdl =
incrementalDynamicKMeans
IsWarm: 0
Metrics: [1×2 table]
NumClusters: 2
NumDynamicClusters: 11
Centroids: [2×0 double]
DynamicCentroids: [11×0 double]
Distance: "sqeuclidean"
Properties, Methods
Mdl is an incrementalDynamicKMeans model object that is prepared for incremental learning.
Fit Incremental Clustering Model
Fit the incremental clustering model Mdl to the data using the fit function. To simulate a data stream, fit the model in chunks of 100 observations at a time. Because WarmupPeriod = 1000, fit only returns cluster indices after the tenth iteration. At each iteration:
Process 100 observations.
Store the number of dynamic clusters in
numDynClusters, to see how it evolves during incremental learning.Overwrite the previous incremental model with a new one fitted to the incoming observations.
Update the simplified silhouette performance metrics (
CumulativeandWindow) using theupdateMetricsfunction.Store the metrics for the merged clusters in
siland the metrics for the dynamic clusters indynsil, to see how they evolve during incremental learning.
numObsPerChunk = 100; n = size(Xtrain,1); nchunk = floor(n/numObsPerChunk); sil = array2table(zeros(nchunk,2),"VariableNames",["Cumulative" "Window"]); dynsil = array2table(zeros(nchunk,2),"VariableNames",["Cumulative" "Window"]); numDynClusters = []; for j = 1:nchunk numDynClusters(j) = Mdl.NumDynamicClusters; ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); chunkrows = ibegin:iend; Mdl = fit(Mdl,Xtrain(chunkrows,:)); Mdl = updateMetrics(Mdl,Xtrain(chunkrows,:)); sil{j,:} = Mdl.Metrics{"SimplifiedSilhouette",:}; dynsil{j,:} = Mdl.DynamicMetrics{"SimplifiedSilhouette",:}; end
Analyze Incremental Model During Training
Plot the number of dynamic clusters at the start of each iteration.
plot(numDynClusters)
xlabel("Iteration");
The model initially has 11 dynamic clusters, and 14 dynamic clusters at the final iteration.
Plot the mean simplified silhouette metric for the merged clusters and the dynamic clusters.
figure; t = tiledlayout(2,1); nexttile h = plot(sil.Variables); ylabel("Simplified Silhouette") xline(Mdl.WarmupPeriod/numObsPerChunk,"b:") legend(h,sil.Properties.VariableNames,Location="southeast") title("Merged Cluster Metrics") nexttile h2 = plot(dynsil.Variables); ylabel("Simplified Silhouette") xline(Mdl.WarmupPeriod/numObsPerChunk,"b:") legend(h2,dynsil.Properties.VariableNames,Location="northeast") xlabel(t,"Iteration") title("Dynamic Cluster Metrics")

After the warm-up period, the updateMetrics function returns performance metrics. A high metric value indicates that, on average, each observation is well matched to its own cluster and poorly matched to other clusters. The higher metric values in the top plot indicate that the merged clusters provide a better clustering solution for the data than the unmerged dynamic clusters.
Analyze the Final Clustering Model Using the Test Set
Create a bar chart of the dynamic cluster counts after the final iteration.
figure
bar(Mdl.DynamicClusterCounts)
xlabel("Dynamic Cluster Number");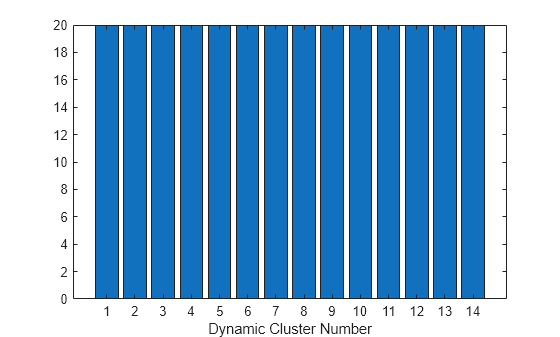
The bar chart shows that the model assigns the observations equally among the dynamic clusters.
Plot the test data set, and color the points according to the dynamic cluster assignments of the final trained model. Plot the dynamic cluster centroids using blue pentagram markers.
C = Mdl.DynamicCentroids; [~,~,dynIdx] = assignClusters(Mdl,Xtest); figure; scatter3(Xtest(:,1),Xtest(:,2),Xtest(:,3),3,dynIdx,"filled"); hold on scatter3(C(:,1),C(:,2),C(:,3),100,"b","Pentagram","filled"); hold off

The dynamic cluster centroids are located within the overall distribution of the observations, and are equally divided among the two groups in the data.
Plot the test data set and color the points according to the merged cluster assignments of the final trained model. Use the color red for the observations whose merged cluster assignments do not match the group identification numbers. Plot the merged cluster centroids using blue pentagram markers.
C = Mdl.Centroids; idx = assignClusters(Mdl,Xtest); incorrectIds = find(idx ~= idsTest); figure; scatter3(Xtest(:,1),Xtest(:,2),Xtest(:,3),1,idx,"filled"); hold on scatter3(C(:,1),C(:,2),C(:,3),100,"b","Pentagram","filled"); scatter3(Xtest(incorrectIds,1),Xtest(incorrectIds,2),Xtest(incorrectIds,3),5,"r","filled") hold off

The plot shows that the merged centroids lie near the center of each group in the data. The observations with incorrect cluster assignments lie mainly in the region in between the two groups.
Use the helper function AdjustedRandIndex to calculate the adjusted Rand index, which measures the similarity of the clustering indices and the group identification numbers.
AdjustedRandIndex(idx,idsTest)
ans = 0.9584
The adjusted Rand index is close to 1, indicating that the clustering model does a good job of correctly predicting the group identification numbers of the test set observations.
function ARI = AdjustedRandIndex(labels1, labels2) % Helper function to calculate the Adjusted Rand Index (ARI) to % measure the similarity between two clustering labels labels1 % and labels2. C = confusionmat(labels1, labels2); n = numel(labels2); % Calculate sums for rows and columns sumRows = sum(C, 2); sumCols = sum(C, 1); ss = sum(C.^2,"all"); TN = ss-n; % True negatives FP = sum(C*sumCols')-ss; % False positives FN = sum(C'*sumRows)-ss; % False negatives TP = n^2-FP-FN-ss; % True positives if FN == 0 && FP == 0 ARI = 1; else ARI = 2*(TP*TN-FN*FP)/((TP+FN)*(FN+TN)+(TP+FP)*(FP+TN)); end end % LocalWords: ARI
Input Arguments
Incremental dynamic k-means clustering model, specified as an
incrementalDynamicKMeans model object. You can create
Mdl by calling incrementalDynamicKMeans
directly.
Chunk of predictor data, specified as a numeric matrix of n
observations and Mdl.NumPredictors variables. The rows of
X correspond to observations, and the columns correspond to
variables. The software ignores observations that contain at least one missing
value.
Note
updateMetrics supports
only numeric input predictor data. If your input data includes categorical data, you
must prepare an encoded version of the categorical data. Use dummyvar to convert each categorical variable to a numeric matrix of
dummy variables. Then, concatenate all dummy variable matrices and any other numeric
predictors. For more details, see Dummy Variables.
Data Types: single | double
Output Arguments
Updated incremental dynamic k-means clustering model, returned as
an incrementalDynamicKMeans model object.
If the input model Mdl is not warm
(Mdl.IsWarm is false),
updateMetrics does not compute performance metrics. As a result,
the Metrics and DynamicMetrics properties of the
output model Mdl contain only NaN values. If the
input model is warm, updateMetrics computes the cumulative and window
performance metrics on the new data X, and overwrites the
corresponding elements of Mdl.Metrics and
Mdl.DynamicMetrics. All other properties of the input model carry
over to the output model. For more details, see Performance Metrics.
More About
When the incremental model is warm (Mdl.IsWarm property) and you pass
new data to the updateMetrics function, the software tracks the model
performance metrics in Mdl.Metrics using
Mdl.Centroids, and in Mdl.DynamicMetrics using
Mdl.DynamicCentroids. An incremental model becomes warm after fit fits the
incremental model to WarmupPeriod observations, which is the
warm-up period. The default performance metric for an incrementalDynamicKMeans model object is
"SimplifiedSilhouette". For more details, see Simplified Silhouette.
If Mdl.EstimationPeriod > 0, the software estimates
hyperparameters before fitting the model to data. Therefore, the software must process an
additional EstimationPeriod observations before the model starts the
warm-up period.
The Metrics and DynamicMetrics properties of the
incremental model each store two forms of each performance metric as variables (columns) of
a table, Cumulative and Window, with individual
metrics in rows. When the incremental model is warm, updateMetrics
updates the metrics at the following frequencies:
Cumulative— The function computes cumulative metrics since the start of model performance tracking. The function updates metrics every time you call it, and bases the calculation on the entire supplied data set until a model reset.Window— The function computes metrics based on all observations within a window determined by theMetricsWindowSizename-value argument.MetricsWindowSizealso determines the frequency at which the software updatesWindowmetrics. For example, ifMetricsWindowSizeis 20, the function computes metrics based on the last 20 observations in the supplied data (X((end – 20 + 1):end,:)andY((end – 20 + 1):end)).Incremental functions that track performance metrics within a window use the following process:
Store
MetricsWindowSizeamount of values for each specified metric.Populate elements of the metrics values with the model performance based on batches of incoming observations.
When the window of observations is filled, overwrite
Mdl.Metrics.WindowandMdl.DynamicMetrics.Windowwith the average performance in the metrics window. If the window is overfilled when the function processes a batch of observations, the latest incomingMetricsWindowSizeobservations are stored, and the earliest observations are removed from the window. For example, supposeMetricsWindowSizeis 20, the window contains 10 stored values from a previously processed batch, and 15 values are incoming. To compose the length 20 window, the functions use the measurements from the 15 incoming observations and the latest 5 measurements from the previous batch.
The software omits an observation with a NaN cluster index when
computing the Cumulative and Window performance metric
values.
The simplified silhouette value si for the ith point is defined as
where ap,i is the distance of
the ith point to the centroid of its cluster p[1].
bp,i is the distance of the
ith point to the centroid of its closest neighboring cluster. If the
ith point is the only point in its cluster, then the simplified
silhouette value of the point is 1.
The simplified silhouette values range from –1 to 1.
A high value indicates that the point is well matched to its own cluster and poorly matched
to other clusters. If most points have a high simplified silhouette value, then the
clustering solution is appropriate. If many points have a low or negative simplified
silhouette value, then the clustering solution might have too many or too few clusters. You
can use simplified silhouette values as a clustering evaluation criterion with any distance
metric. By default, the performance metric values stored in the model object are the average
simplified silhouette values for all points passed to the updateMetrics
function.
References
[1] Vendramin, Lucas, Ricardo J.G.B. Campello, and Eduardo R. Hruschka. On the Comparison of Relative Clustering Validity Criteria. In Proceedings of the 2009 SIAM international conference on data mining, 733–744. Society for Industrial and Applied Mathematics, 2009.
Version History
Introduced in R2025a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)