plotResidualDistribution
Plot the distribution of the residuals
Description
plotResidualDistribution(
shows the normal probability plot and the corresponding histogram of the residuals.
Use the plot to check if the distribution of the residuals deviates from the
normality.resultsObj)
Examples
Load the sample data set.
load data10_32R.mat gData = groupedData(data); gData.Properties.VariableUnits = ["","hour","milligram/liter","milligram/liter"];
Create a two-compartment PK model.
pkmd = PKModelDesign; pkc1 = addCompartment(pkmd,"Central"); pkc1.DosingType = "Infusion"; pkc1.EliminationType = "linear-clearance"; pkc1.HasResponseVariable = true; pkc2 = addCompartment(pkmd,"Peripheral"); model = construct(pkmd); configset = getconfigset(model); configset.CompileOptions.UnitConversion = true; responseMap = ["Drug_Central = CentralConc","Drug_Peripheral = PeripheralConc"];
Provide model parameters to estimate.
paramsToEstimate = ["log(Central)","log(Peripheral)","Q12","Cl_Central"]; estimatedParam = estimatedInfo(paramsToEstimate,'InitialValue',[1 1 1 1]);
Assume every individual receives an infusion dose at time = 0, with a total infusion amount of 100 mg at a rate of 50 mg/hour.
dose = sbiodose("dose","TargetName","Drug_Central"); dose.StartTime = 0; dose.Amount = 100; dose.Rate = 50; dose.AmountUnits = "milligram"; dose.TimeUnits = "hour"; dose.RateUnits = "milligram/hour";
Estimate model parameters. By default, the function estimates a set of parameter for each individual (unpooled fit).
fitResults = sbiofit(model,gData,responseMap,estimatedParam,dose);
Plot the results.
plot(fitResults);
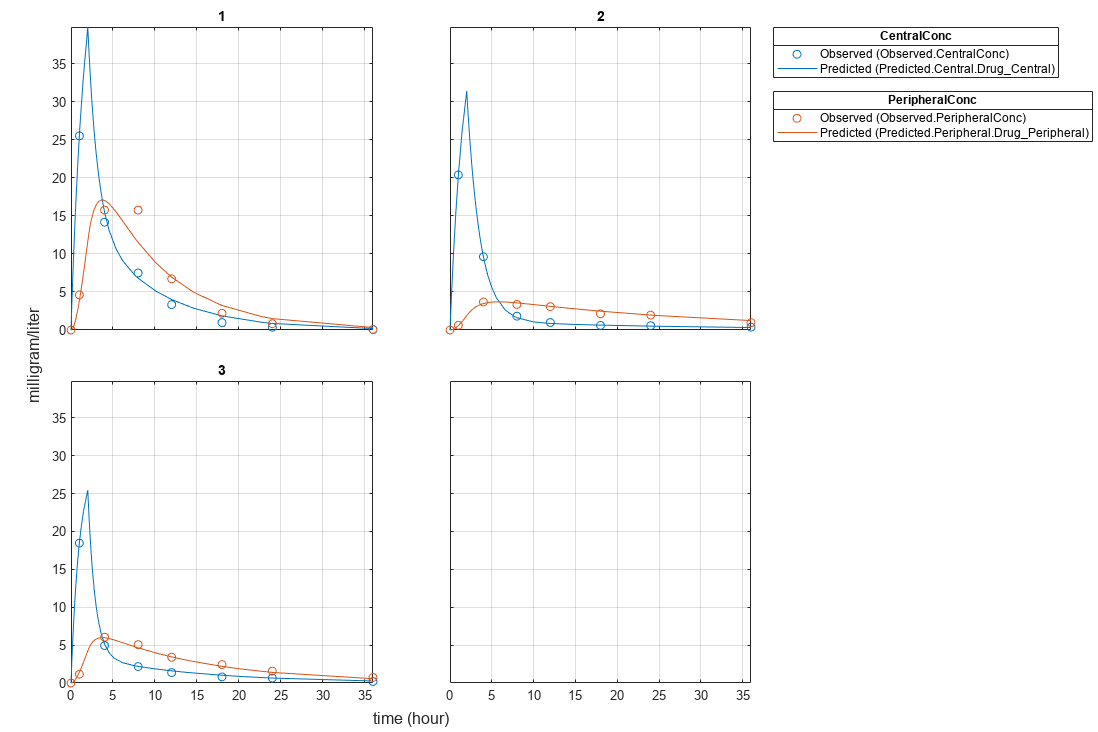
Plot all groups in one plot.
plot(fitResults,"PlotStyle","one axes");
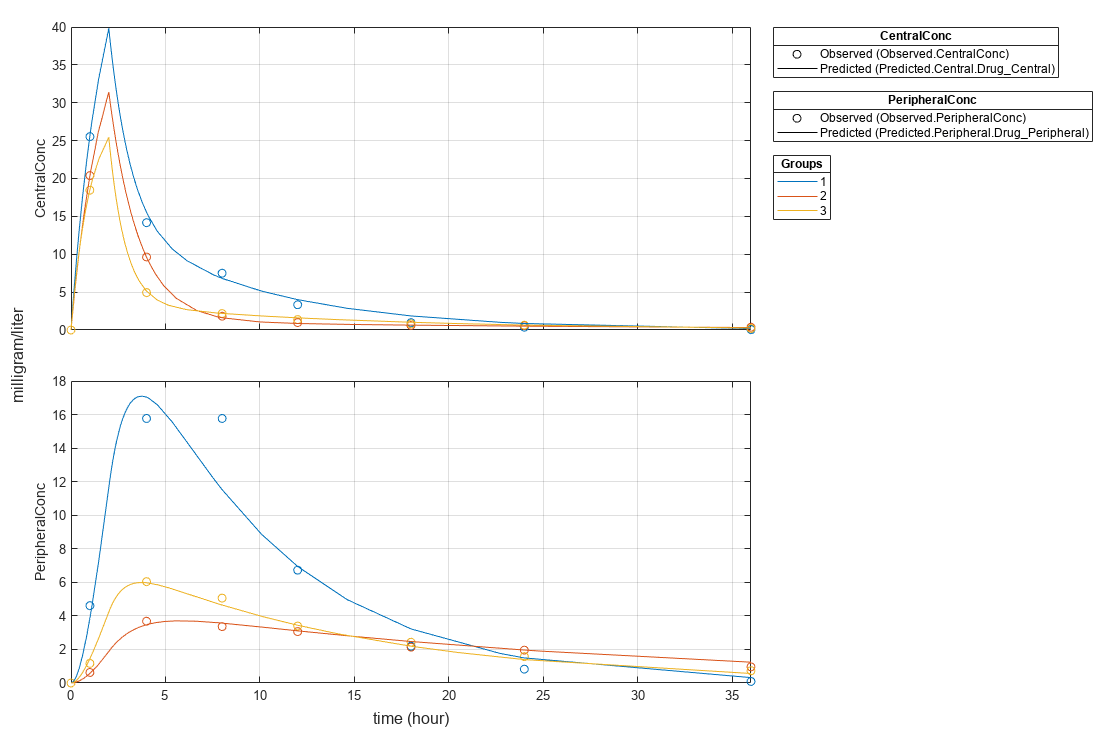
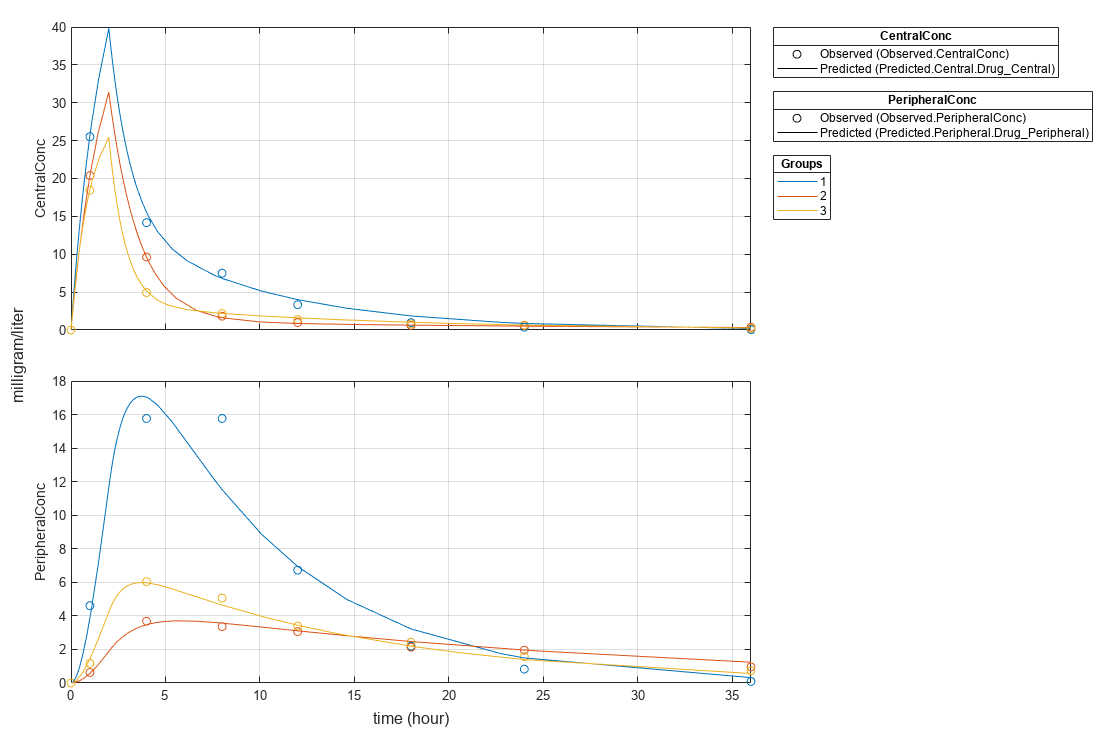
Change some axes properties.
s = struct; s.Properties.XGrid = "on"; s.Properties.YGrid = "on"; plot(fitResults,"PlotStyle","one axes","AxesStyle",s);
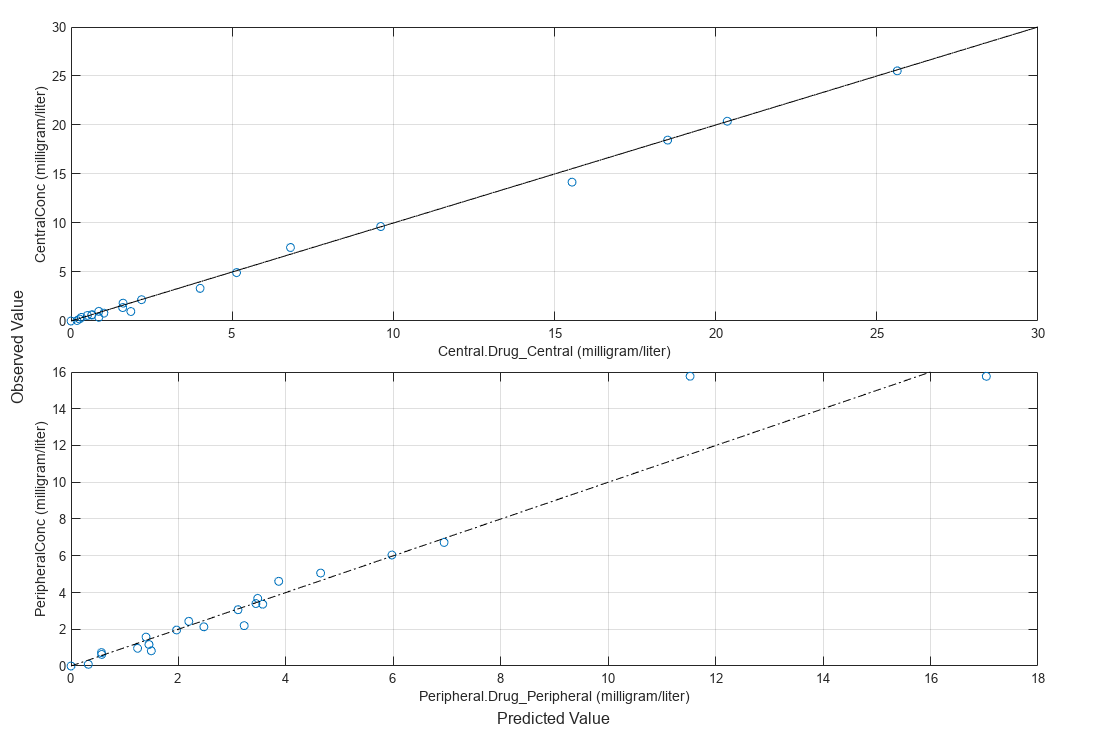
Compare the model predictions to the actual data.
plotActualVersusPredicted(fitResults)
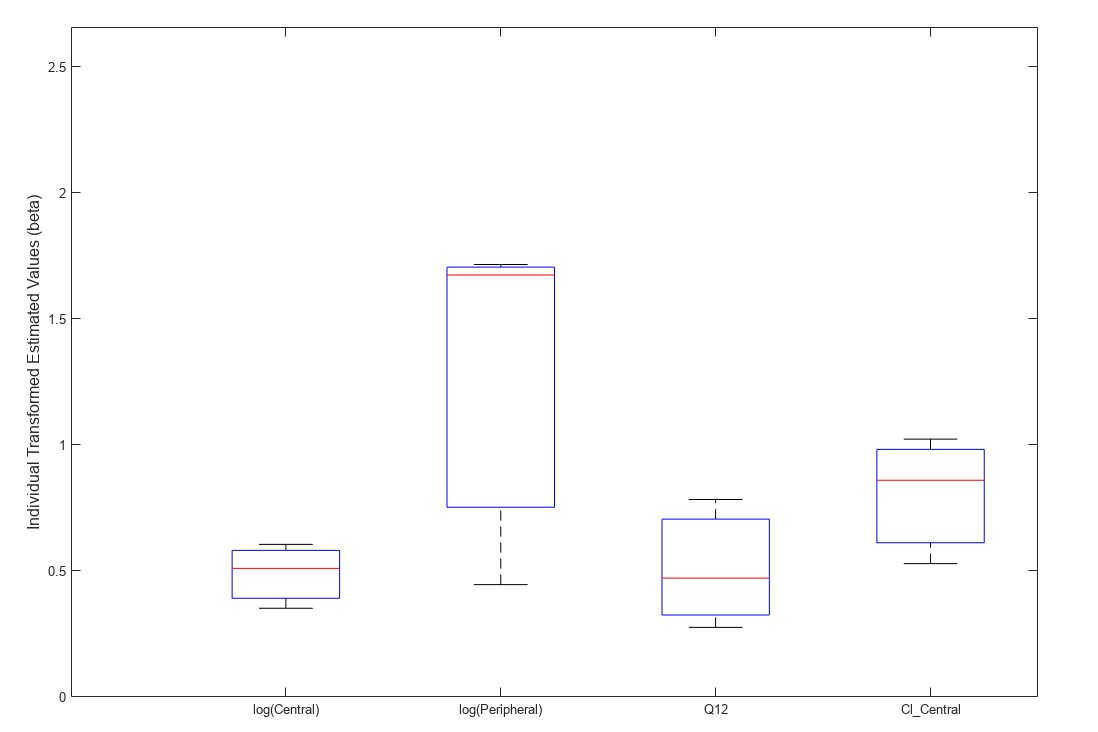
Use boxplot to show the variation of estimated model parameters.
boxplot(fitResults)
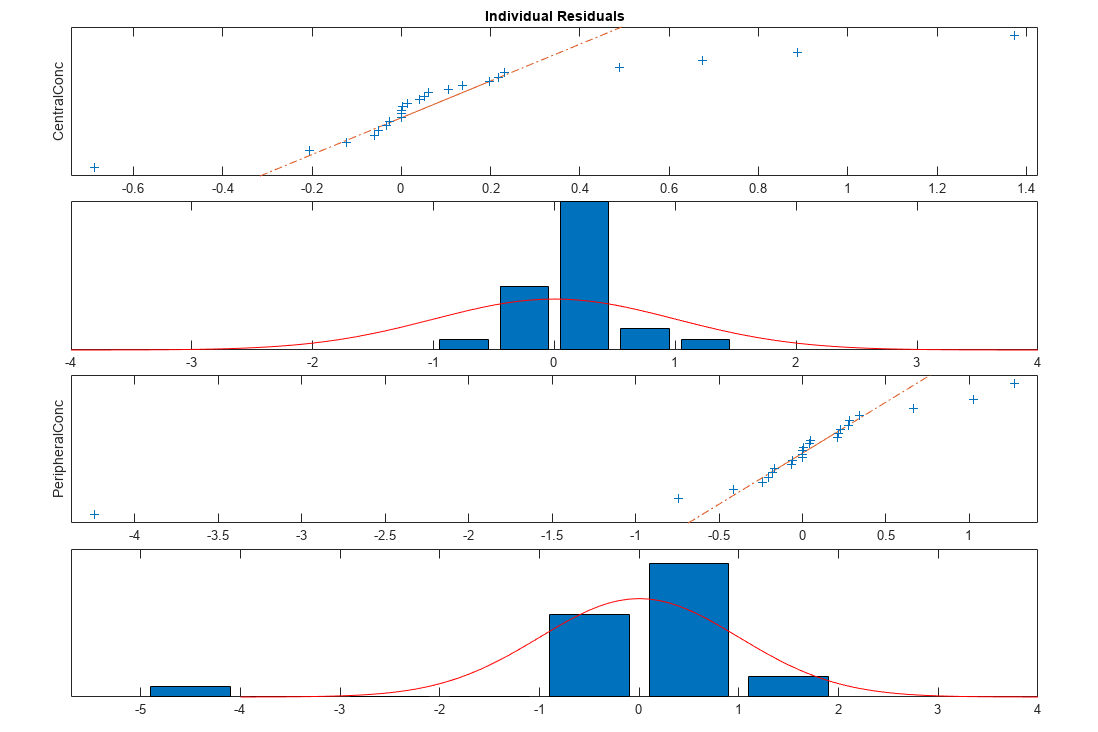
Plot the distribution of residuals. This normal probability plot shows the deviation from normality and the skewness on the right tail of the distribution of residuals. The default (constant) error model might not be the correct assumption for the data being fitted.
plotResidualDistribution(fitResults)
Plot residuals for each response using the model predictions on x-axis.
plotResiduals(fitResults,"Predictions")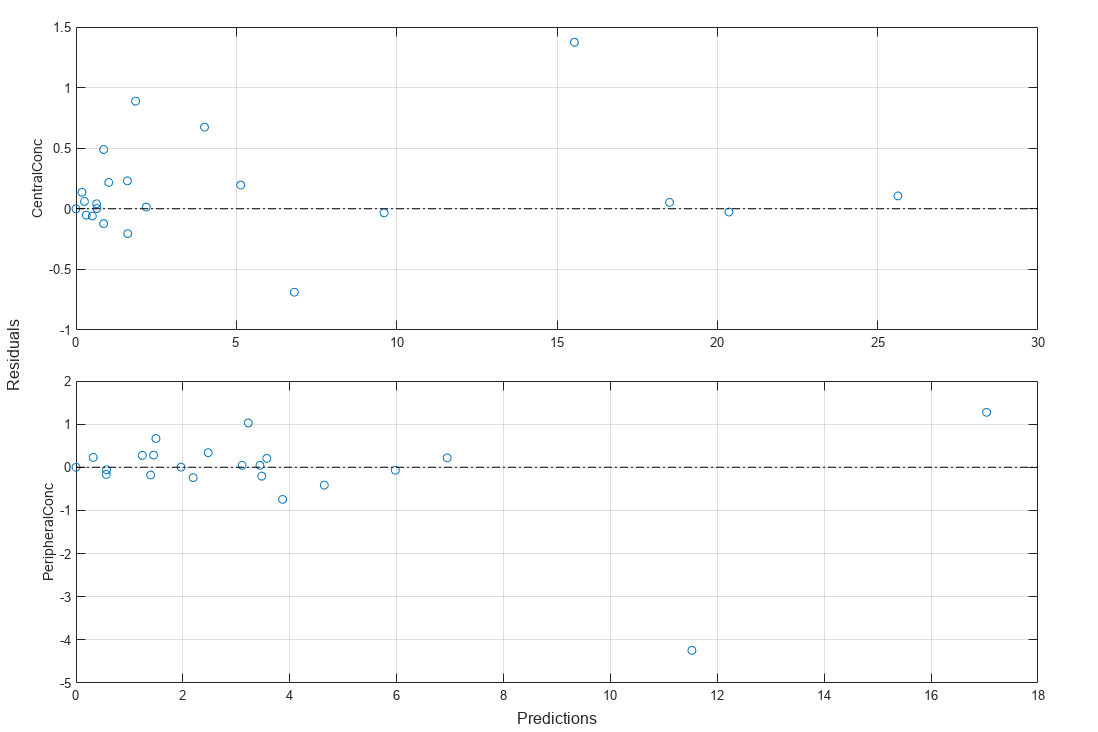
Get the summary of the fit results. stats.Name contains the name for each table from stats.Table, which contains a list of tables with estimated parameter values and fit quality statistics.
stats = summary(fitResults); stats.Name
ans = 'Unpooled Parameter Estimates'
ans = 'Statistics'
ans = 'Unpooled Beta'
ans = 'Residuals'
ans = 'Covariance Matrix'
ans = 'Error Model'
stats.Table
ans=3×9 table
Group Central Estimate Central StandardError Peripheral Estimate Peripheral StandardError Q12 Estimate Q12 StandardError Cl_Central Estimate Cl_Central StandardError
_____ ________________ _____________________ ___________________ ________________________ ____________ _________________ ___________________ ________________________
{'1'} 1.422 0.12334 1.5619 0.36355 0.47163 0.15196 0.5291 0.036978
{'2'} 1.8322 0.019672 5.3364 0.65327 0.2764 0.030799 0.86035 0.026257
{'3'} 1.6657 0.038529 5.5632 0.37063 0.78361 0.058657 1.0233 0.027311
ans=3×7 table
Group AIC BIC LogLikelihood DFE MSE SSE
_____ _______ _______ _____________ ___ ________ _______
{'1'} 60.961 64.051 -26.48 12 2.138 25.656
{'2'} -7.8379 -4.7475 7.9189 12 0.029012 0.34814
{'3'} -1.4336 1.6567 4.7168 12 0.043292 0.5195
ans=3×9 table
Group Central Estimate Central StandardError Peripheral Estimate Peripheral StandardError Q12 Estimate Q12 StandardError Cl_Central Estimate Cl_Central StandardError
_____ ________________ _____________________ ___________________ ________________________ ____________ _________________ ___________________ ________________________
{'1'} 0.35208 0.086736 0.44589 0.23277 0.47163 0.15196 0.5291 0.036978
{'2'} 0.60551 0.010737 1.6746 0.12242 0.2764 0.030799 0.86035 0.026257
{'3'} 0.51027 0.02313 1.7162 0.066621 0.78361 0.058657 1.0233 0.027311
ans=24×4 table
ID Time CentralConc PeripheralConc
__ ____ ___________ ______________
1 0 0 0
1 1 0.10646 -0.74394
1 4 1.3745 1.2726
1 8 -0.68825 -4.2435
1 12 0.67383 0.21806
1 18 0.88823 1.0269
1 24 0.48941 0.66755
1 36 0.13632 0.22948
2 0 0 0
2 1 -0.026731 -0.058311
2 4 -0.033299 -0.20544
2 8 -0.20466 0.20696
2 12 -0.12223 0.045409
2 18 0.041224 0.33883
2 24 -0.059498 0.0036257
2 36 -0.051645 0.27616
⋮
ans=12×6 table
Group Parameters Central Peripheral Q12 Cl_Central
_____ ______________ ___________ __________ ___________ ___________
{'1'} {'Central' } 0.015213 -0.022539 -0.0086672 0.001159
{'1'} {'Peripheral'} -0.022539 0.13217 0.045746 -0.0073135
{'1'} {'Q12' } -0.0086672 0.045746 0.023092 -0.0021484
{'1'} {'Cl_Central'} 0.001159 -0.0073135 -0.0021484 0.0013674
{'2'} {'Central' } 0.00038701 -0.002161 -0.00010177 9.7448e-05
{'2'} {'Peripheral'} -0.002161 0.42676 0.019101 -0.015755
{'2'} {'Q12' } -0.00010177 0.019101 0.00094857 -0.00073328
{'2'} {'Cl_Central'} 9.7448e-05 -0.015755 -0.00073328 0.00068942
{'3'} {'Central' } 0.0014845 -0.0054648 -0.0013216 0.00016639
{'3'} {'Peripheral'} -0.0054648 0.13737 0.016903 -0.0072722
{'3'} {'Q12' } -0.0013216 0.016903 0.0034406 -0.00082538
{'3'} {'Cl_Central'} 0.00016639 -0.0072722 -0.00082538 0.00074587
ans=3×5 table
Group Response ErrorModel a b
_____ __________ ____________ _______ ___
{'1'} {0×0 char} {'constant'} 1.2663 NaN
{'2'} {0×0 char} {'constant'} 0.14751 NaN
{'3'} {0×0 char} {'constant'} 0.18019 NaN
Input Arguments
Estimation results, specified as an OptimResults object or
NLINResults object, or
vector of results objects which contains estimation results from running
sbiofit.
Version History
Introduced in R2014a
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)