seqqcplot
Create quality control plots for sequence and quality data in MATLAB
Syntax
Description
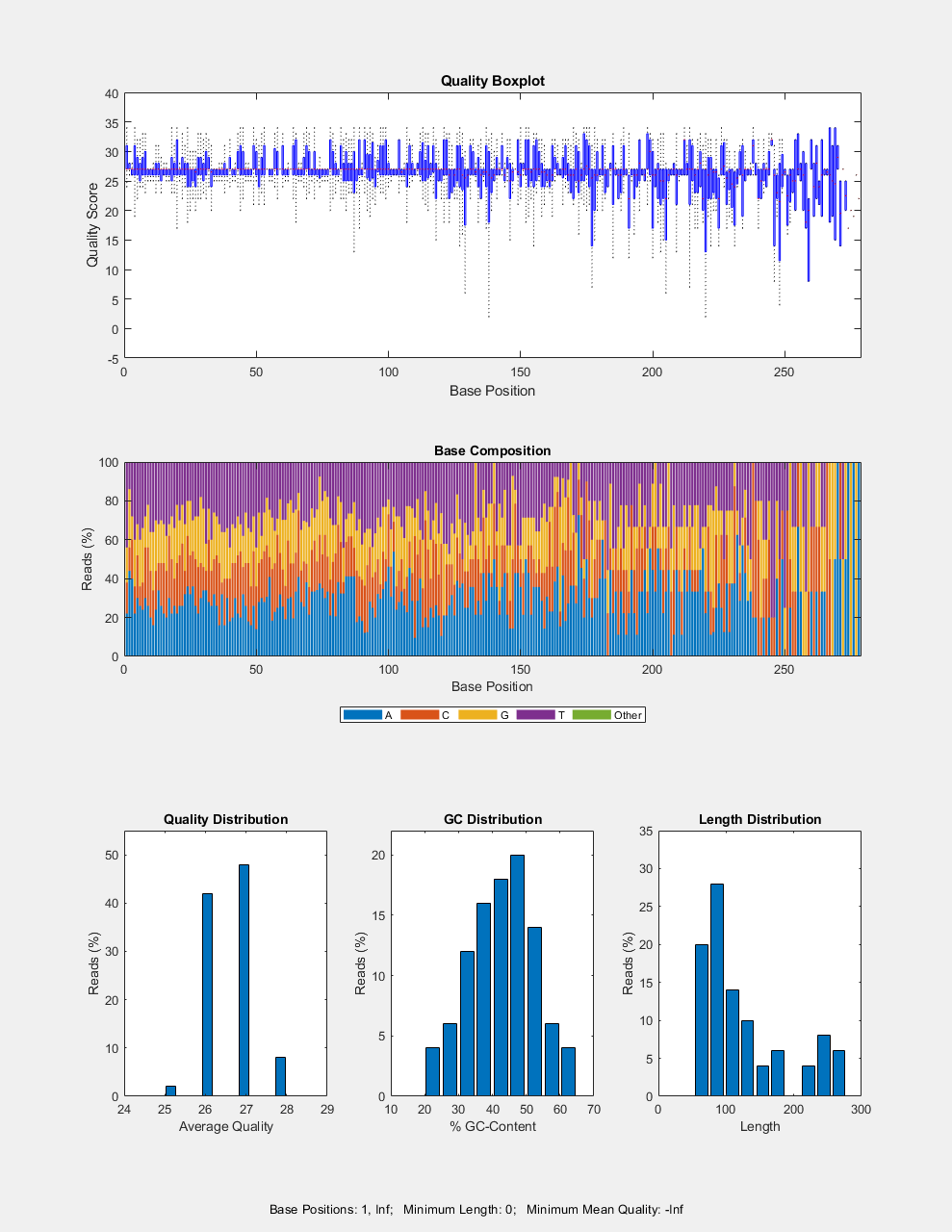
seqqcplot( generates a figure with
quality control (QC) plots of sequence and quality data from
dataSource)dataSource in MATLAB. The figure contains the following
types of QC plots.
Box plot for the average quality score at each sequence position
Bar plot for the sequence base composition at each sequence position
Histogram of the average sequence quality score distribution
Histogram of the GC-content distribution
Histogram of the sequence length distribution
In the figure, you can click a specific plot to open it in a separate window.
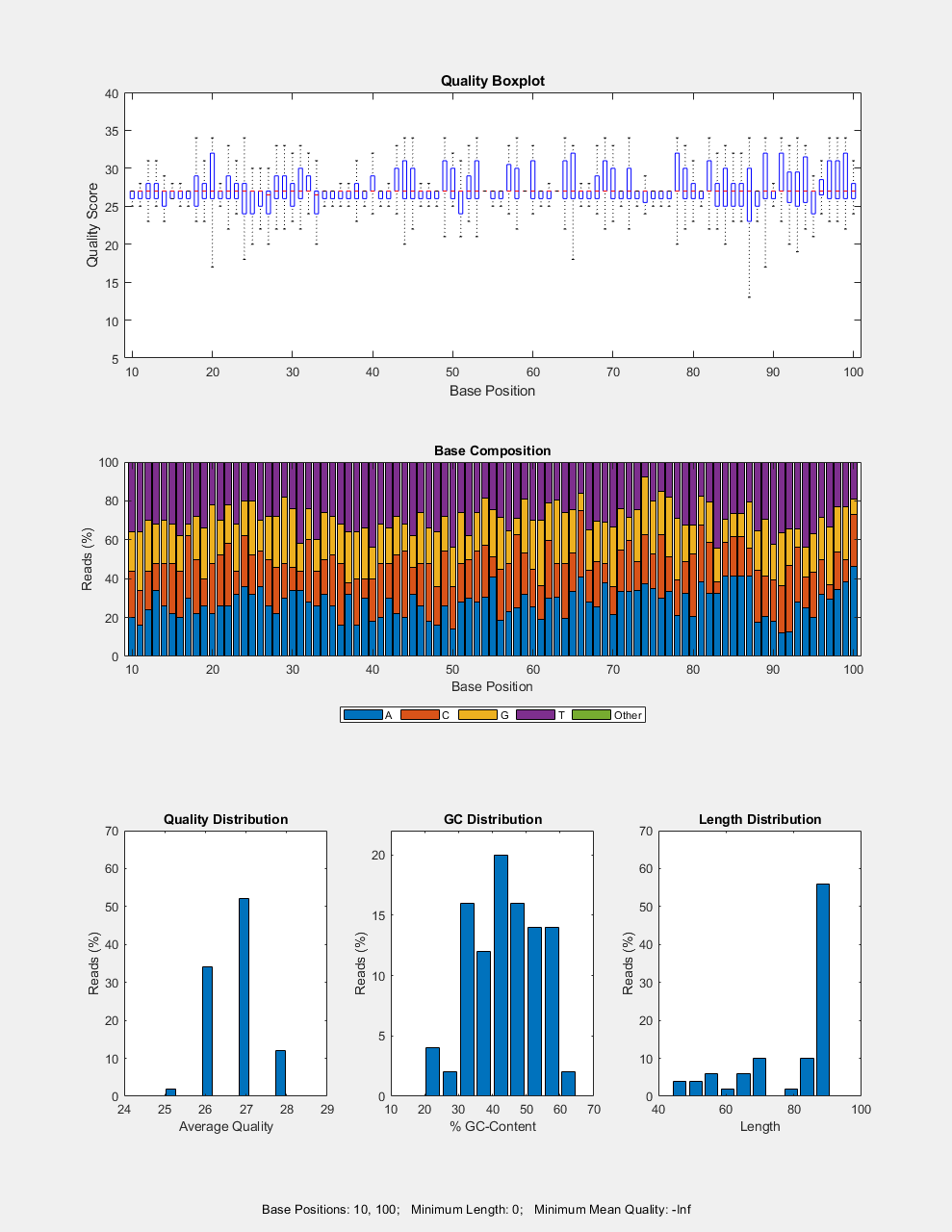
seqqcplot(
generates a QC plot specified by dataSource,type)type.
seqqcplot(
also specifies the encoding format of the base quality in the input file.dataSource,type,encoding)
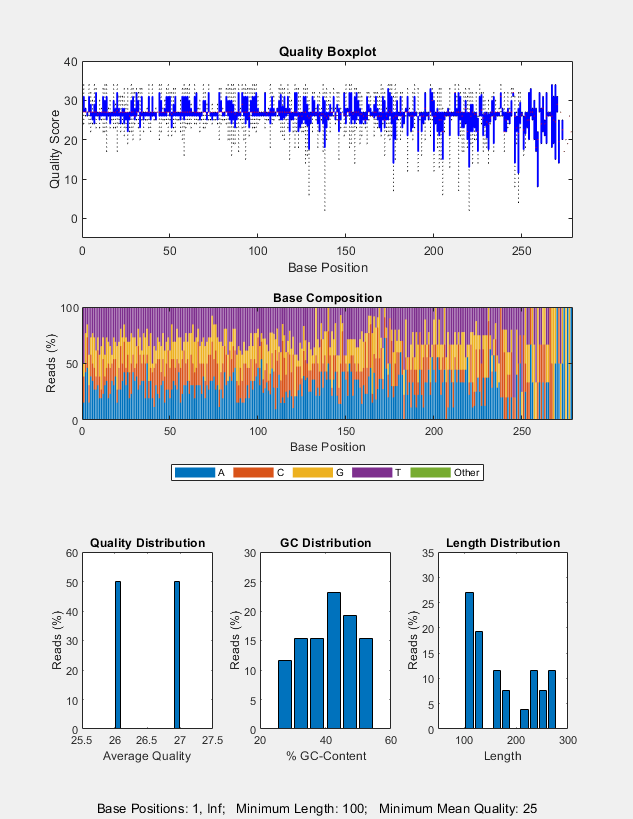
seqqcplot(___, uses
any of the input arguments in the previous syntaxes and additional
options specified by one or more Name,Value)Name,Value pair
arguments.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Version History
Introduced in R2017a
See Also
seqtrim | seqfilter | seqsplit | seqsplitpe