seqlinkage
Construct phylogenetic tree from pairwise distances
Syntax
Description
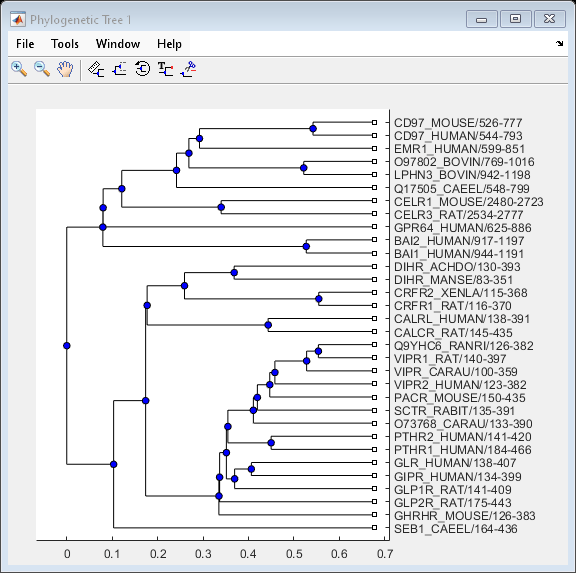
Examples
Input Arguments
Name-Value Arguments
Version History
Introduced before R2006aSee Also
phytree | phytreewrite | seqpdist | seqneighjoin | cluster | plot | view