sbioncaoptions
Specify options to calculate noncompartmental analysis (NCA) parameters
Syntax
Description
Examples
Load a synthetic data set that contains the drug concentration measurements of four individuals after an IV bolus dose.
load data1.matSet the dose amounts to NaN at time points when no dose was administered.
data1.Dose(data1.Dose(:) == 0) = NaN;
Display the data.
sbiotrellis(data1,'ID','Time','DrugConc','Marker','o','LineStyle','--');
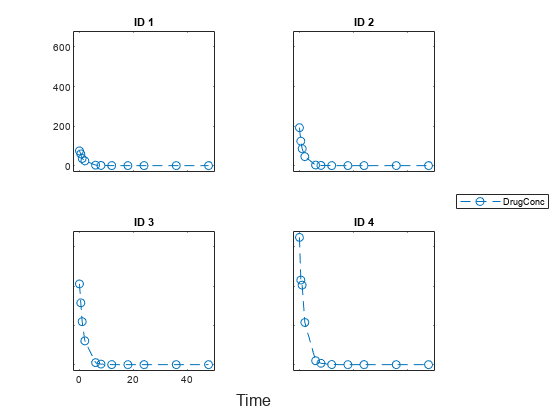
Categorize the data columns using an NCA options object.
opt = sbioncaoptions; opt.groupColumnName = 'ID'; opt.concentrationColumnName = 'DrugConc'; opt.timeColumnName = 'Time'; opt.IVDoseColumnName = 'Dose';
Compute NCA parameters for each individual.
ncaparameters = sbionca(data1,opt);
Display the first few columns of the table. Each row of ncaparameters table represents an individual (or group), and each column lists the corresponding NCA parameter value.
ncaparameters(:,1:15)
ans=4×15 table
ID doseSchedule administrationRoute Lambda_Z R2 adjusted_R2 Num_points AUC_0_last Tlast C_max C_max_Dose T_max MRT T_half AUC_infinity
__ ____________ ___________________ ________ _______ ___________ __________ __________ _____ ______ __________ _____ ______ ______ ____________
1 {'Single'} {'IVBolus'} 0.57893 0.99991 0.9999 11 143.61 48 74.412 1.4882 0 1.5408 1.1973 143.61
2 {'Single'} {'IVBolus'} 0.66798 0.99998 0.99998 11 299.37 48 191.96 1.9196 0 1.3352 1.0377 299.37
3 {'Single'} {'IVBolus'} 0.62124 0.99999 0.99999 11 766.5 48 411.06 1.6442 0 1.4476 1.1157 766.5
4 {'Single'} {'IVBolus'} 0.58011 0.99995 0.99995 11 1301.8 48 648.33 1.2967 0 1.5721 1.1949 1301.8
You can also specify a custom time range to compute T_max and C_max within that time range, say from time = 0 to 20. You can do so by setting the C_max_ranges property as a cell array of two-element row vector.
opt.C_max_ranges = {[5.5 20]};
ncaparameters2 = sbionca(data1,opt);The function reports the T_max and C_max values within the range by adding two new columns: T_max_5_5__20 and C_max_5_5__20. Note that in the names of these two columns, the last time point is preceded by two consecutive underscores (__).
ncaparameters2.T_max_5_5__20(:)
ans = 4×1
6
6
6
6
ncaparameters2.C_max_5_5__20(:)
ans = 4×1
2.2719
3.0213
10.0233
19.9006
Similarly, you can specify a custom time range to compute the partial AUC value for each group.
opt.PartialAreas = {[0 20]};
ncaparameters3 = sbionca(data1,opt);
ncaparameters3.AUC_0__20(:)ans = 4×1
103 ×
0.1436
0.2994
0.7665
1.3017
You can also specify multiple time ranges for C_max_ranges and PartialAreas.
opt.C_max_ranges = {[0 20],[0 10],[0 15]};
opt.PartialAreas = {[0 12],[0 30]};
ncaparameters4 = sbionca(data1,opt);Output Arguments
Options to calculate NCA parameters, returned as a
SimBiology.nca.Options object. The properties of the
object are classified into two groups, data classification options and
parameter calculation options.
Data Classification Options
| Property | Description |
|---|---|
IVDoseColumnName | Name of the data column that contains the IV dose amount. |
EVDoseColumnName | Name of the data column that contains the extravascular (EV) dose amount. |
concentrationColumnName | Name of the data column that contains the measured concentrations. |
timeColumnName | Name of the data column that contains the time points. |
groupColumnName |
Name of the data column that contains the grouping
information. You can specify grouping using two
levels of hierarchy. Specify the outer level of
grouping in this column. Specify the inner level of
grouping (subgroups) in
If you specify For example, consider data that contains three groups, where each group contains four patients. The group column labels the three groups, and the ID column labels each patient. |
idColumnName |
Name of the data column that contains the grouping
information. You can specify grouping using two
levels of hierarchy. Specify the inner level of
grouping (subgroups) in this column. Specify the
outer level of grouping in
If you specify |
infusionRateColumnName | Name of the data column that contains the infusion rates. |
Parameter Calculation Options
| Property | Description |
|---|---|
AUCCalculationMode (since R2024a) | Method to calculate the AUC. There are two methods:
|
LOQ | Lower limit of quantization, a threshold below which the values of dependent variable are truncated to zero. |
AdministrationRoute | Drug administration route. Three types of
administration are supported:
IVBolus,
IVInfusion, and
ExtraVascular. |
TAU | Dosing interval for multiple-dosing data. |
SparseData | Boolean that indicates whether or not the values of dependent variable are averaged between subgroups to further populate a profile for a group. Time values for each measurement across subgroups (IDs) within a group must be identical. |
Lambda_Z_Time_Min_Max |
Two-element row vector that specifies a custom time range to compute the terminal rate constant (Lambda_z). The time range applies to all groups; you cannot specify a different time range for each group. For details, see Noncompartmental Analysis. |
PartialAreas |
Cell array of one or more two-element row vectors that specify one or more time ranges used to compute the partial AUC values. You can specify multiple rows for group-specific ranges, where the number of rows equal the number of groups. If there is only one row, the same time ranges are used for all groups. |
C_max_ranges |
Cell array of one or more two-element row vectors
that specify one or more time ranges used to report
the |
More About
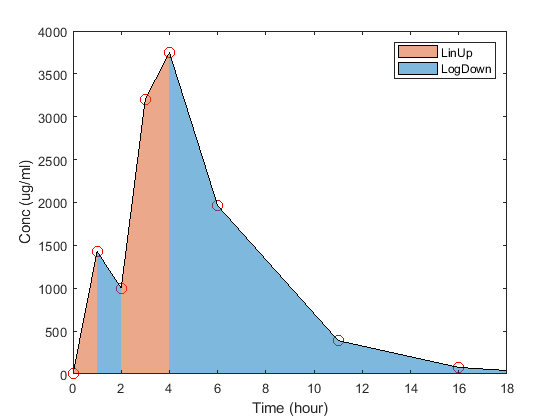
This method uses the linear trapezoidal integration
(LinUp) whenever concentrations are increasing or constant,
and trapezoidal integration of the log-transformed concentrations
(LogDown) whenever they are decreasing.
For the LinUpLogDown method, SimBiology uses the following equation to calculate the AUC for log-transformed concentrations:
where, C1 and C2 are concentrations at times t1 and t2, respectively. ΔT is t2 – t1.
For instance, the next figure illustrates where LinUp and
LogDown are used for a given concentration-time course
depending on whether concentrations are decreasing or increasing.
Version History
Introduced in R2017b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Website auswählen
Wählen Sie eine Website aus, um übersetzte Inhalte (sofern verfügbar) sowie lokale Veranstaltungen und Angebote anzuzeigen. Auf der Grundlage Ihres Standorts empfehlen wir Ihnen die folgende Auswahl: .
Sie können auch eine Website aus der folgenden Liste auswählen:
So erhalten Sie die bestmögliche Leistung auf der Website
Wählen Sie für die bestmögliche Website-Leistung die Website für China (auf Chinesisch oder Englisch). Andere landesspezifische Websites von MathWorks sind für Besuche von Ihrem Standort aus nicht optimiert.
Amerika
- América Latina (Español)
- Canada (English)
- United States (English)
Europa
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)