Human Activity Recognition Using ESP32 Board and LSM9DS1 IMU Sensor
This example shows how to classify and predict one of three different physical human activities: Sitting, Standing and Walking based on data acquired using an ESP32 board and an lsm9ds1 sensor. This example uses Classification Learner (Statistics and Machine Learning Toolbox) app to train a machine learning model with the sensor data. To access all the files for this example, click Open Live Script and download the attached files.
Required Hardware
ESP32-WROOM-DevKitV1
LSM9DS1 IMU sensor
Required MathWorks® Products
MATLAB®
Statistics and Machine Learning Toolbox™
Signal Processing Toolbox
MATLAB Support Package for Arduino Hardware
Hardware Setup
Configure ESP32 hardware for WiFi based workflow. For more information, see Connection over Wi-Fi.
Connect SCL and SDA of ESP32 to SCL and SDA of LSM9DS1 IMU.
Connect the ground and 3.3V pin of ESP32 to ground and VDD of LSM9DS1 IMU.
Place the entire setup in a pocket with the sensor facing away from you.
Capture Dataset
You can either use the three data sets, Sitting_raw_data.mat, Standing_raw_data.mat and Walking_raw_data.mat corresponding to the three different types of physical activities: Sitting, Standing and Walking, provided with this example or capture the data manually.
To use the three data set, click Open Live Script and download Sitting_raw_data.mat, Standing_raw_data.mat and Walking_raw_data.mat files.
To capture the data manually, place the setup in your pocket with the sensor facing away from you. Click the checkbox for iscollectData and start capturing the data. For this example, the data set contains 3200 samples, as defined by the variable noOfSamples, for each of the three physical human activities. Once the prompt '*** Collecting data for activity = ' Sitting ' ***'' appears on the screen, start capturing data for the Sitting activity. Continue capturing the Sitting data until a new prompt appears, asking you to capture the data for Standing and Walking activities, respectively. At the end of data collection, you will have Sitting_raw_data.mat, Standing_raw_data.mat and Walking_raw_data.mat dataset corresponding to the three different types of physical activities.
% Check this if you want to manually collect the data iscollectData =false; % Create activity list activityList = {'Sitting' , 'Standing', 'Walking'}; % Collect data for all activities noOfSamples = 3200; if iscollectData for idx = 1:length(activityList) collectData(activityList{idx},noOfSamples); end end
Feature Extraction
Choose a buffer size based on the required granularity of the extracted features. For this example, set bufferSize to 32.
The extractFeatures function is used to extract features from the data acquired using the accelerometer on the LSM9DS1 IMU sensor. The extracted features are stored in the variable features. To use the extractFeatures function, click Open Live Script and download the extractFeatures.m file.
The activityIDList contains the IDs corresponding to the physical activities where activity ID 1 corresponds to Sitting, 2 corresponds to Standing, and 3 corresponds to Walking.
% Initialize buffer size bufferSize = 32; features =[]; activityIDList = []; % Extract features for idx = 1: length(activityList) data = load([activityList{idx} '_raw_data.mat'],'data'); data = data.data; % Calculate the number of groups numGroups = floor(size(data,1) / bufferSize); for i = 1:numGroups startIndex = (i-1) * bufferSize + 1; endIndex = i * bufferSize; xBuffer = data(startIndex:endIndex,1); yBuffer = data(startIndex:endIndex,2); zBuffer = data(startIndex:endIndex,3); features = [features; extractFeatures(xBuffer, yBuffer, zBuffer)]; end activityIDList = [activityIDList;ones(numGroups,1)*idx]; end
Model Training
Use the Classification Learner app to train a classification model. To open the Classification Learner app, type classificationLearner at the MATLAB command line.
Alternatively, in the Apps tab click the arrow at the right of the Apps section to open the gallery. Then, under Machine Learning and Deep Learning, click Classification Learner.
In the app, click the New Session button, and then click From Workspace.
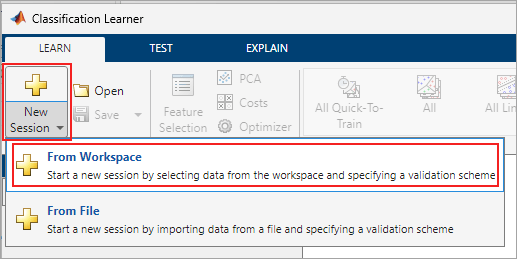
In the Data set pane, click the arrow under Data Set Variable, and then select features. Select From Workspace under Response and select activityIDList. The default option or the Validation Scheme is 5-fold cross-validation, which protects against over-fitting.
Click Start Session.
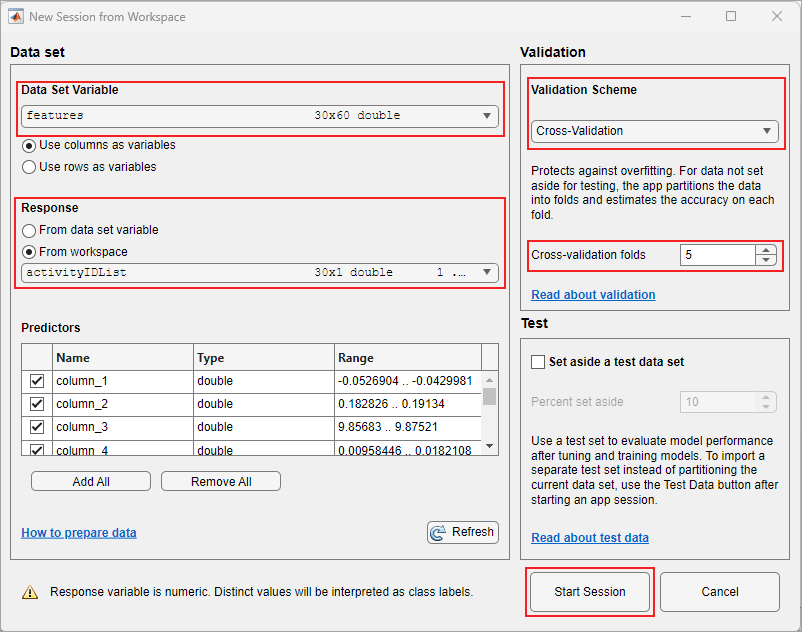
Classification Learner app loads the data set and plots a scatter plot of the first two features.

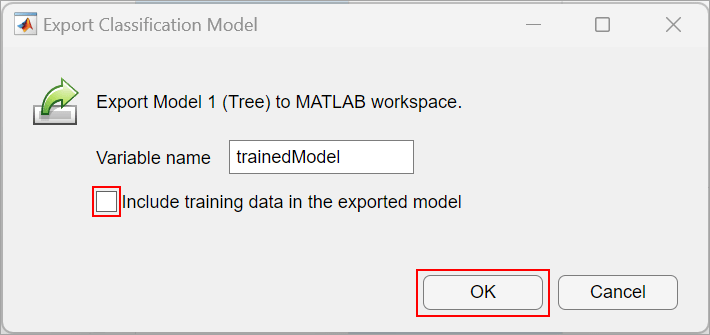
On the app toolstrip, in the Train section, click Train All and select Train Selected. When the training is complete, the Models pane displays the 5-fold, cross-validated classification accuracy. For this example the accuracy is 99.3%, corresponding to the data set provided with this example.
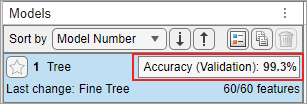
To see the Validation Confusion Matrix, in the Plots and Results section of the app toolstrip, click Confusion Matrix (Validation).
On the app toolstrip, in the Export section, and then click Export Model.
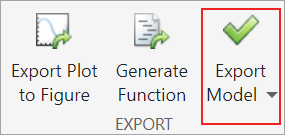
In the Export Classification Model dialog box, clear the check box to exclude the training data and export a compact model, and then click OK.
The structure trainedModel appears in the MATLAB workspace. Save the model using the following command.
save('trainedModel.mat','trainedModel');
Prediction
Run this MATLAB code to predict the activity.
% Create an arduino object for ESP32 board over WiFi aObj = arduino; % Create IMU sensor object imuObj = lsm9ds1(aObj); % Predict the activity for three minutes tic; while(toc < 180) % Initialize buffer and buffer size xBuffer = []; yBuffer = []; zBuffer = []; bufferSize = 32; % Get buffered sensor values for idx = 1:bufferSize % Read the sensor values and calibrate accelerationValues = readAcceleration(imuObj); % Calibrate the acceleration values xBuffer(end+1) = accelerationValues(1); yBuffer(end+1) = accelerationValues(2); zBuffer(end+1) = accelerationValues(3); end % Extract features features = extractFeatures(xBuffer', yBuffer', zBuffer'); % Predict activity y = predictActivity(features); disp(['Detected activity = ' activityList{y}]); end
Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting Detected activity = Sitting